BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://journal.zums.ac.ir/article-1-6637-en.html
2- Dept. of Microbiology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran ,
3- Dept. of Microbiology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
✅ In this study, aminoglycoside-resistant A. baumannii was found in a high percentage of ICU patients, mainly with the enzyme-modified aminoglycosides like aac(6′)-Ib, aph(2'')-Id and ant(3'')-I. ERIC-PCR has also shown an increased level of diversity in A. baumannii isolates. Therefore, genetic diversity or clonal relatedness of A. baumannii isolates in clinical settings can be assessed using ERIC-PCR.
Due to limited therapeutic options, Acinetobacter baumannii accounts for a high mortality rate induced by opportunistic nosocomial infections. A. baumanni-induced infections include meningitis, wounds, ventilator- associated pneumonia, peritonitis, bacteremia, and soft-tissue infections (1, 2). Nosocomial infections resulting from Acinetobacter baumannii are an increasing concern around the world, as they are resistant to many antimicrobial agents and can survive in different hospital environments (2, 3). Due to MDR, at least three groups of antibiotics consisting of quinolones, beta-lactams, carbapenems, and aminoglycosides are ineffective on A. baumannii. Therefore, healthcare systems in Iran are facing a significant challenge (2, 4). Additional studies have reported various pan drug-resistant (PDR) A. baumannii strains (5, 6). Limited therapeutic options have made treating MDR A. baumannii strain infections difficult (2). To fight A. baumannii infections in hospitalized patients, aminoglycosides, whether taken alone or in combination with β-lactams, have been used and remain an essential tool in the fight against MDR strain infections(7). The specific interaction of antimicrobial agents with the 30S ribosomal subunit components, 16S rRNA, blocks translation initiation (8). Aminoglycoside- modifying enzymes (AMEs) such as aminoglycoside O-nucleotidyltransferases (ANTs) (5, 9), aminoglycoside N-acetyltransferases (AACs), and aminoglycoside O-phosphotransferases (APHs) in A. baumanni cause aminoglycoside resistance. APHs are correlated with increased gentamicin resistance (5). A. baumannii has been reported to have a variety of AMEs, consisting of phosphotransferase variants: APH (3')-I, APH (3')-II, and APH(2'')-Ib, as well as variants of the nucleotidyltransferases ANT(3)-I, ANT(4')-I, ANT(2'')-I and the acetyltransferases AAC(3)-I, AAC(6')-I, AAC(6')-II, and AAC(6')-III(10). In addition, methylation of 16S rRNA contributes to the inactivation of aminoglycosides (8). Hyper resistance to aminoglycosides, except streptomycin, is caused by ArmA, RmtA, RmtB, RmtC, and RmtD (8،9). Acinetobacter baumannii usually acquires aminoglycoside resistance genes via transposons, plasmids, or class 1 integrons (Int‑1) containing single or multiple gene cassettes (5, 6). Thus, these resistance genes can be transmitted to other bacterial species.
The genotyping of A. baumannii is an essential method for the specification of the connection between genetics and epidemiology. ERIC-PCR (Enterobacterial repetitive intergenic consensus polymerase chain reaction) is a reliable and fast method for determining the genetic diversity or clonality of A. baumannii isolates (11, 12).
We examined the aminoglycoside resistance genes (aph (2'')-Id, ant (4′)-Ia, ant (3'')-I, aac (6′)-Ib, aac (3)-I, aph (3′)-I, aph (2'')-Ib, aph (2'')-Ic). We used ERIC-PCR to molecularly type MDR A. baumannii gathered from ICU patients with an immune deficiency.
baumannii samples
A prior study obtained 100 clinical isolates from urine, chest, wound swabs, blood, tube secretions, sputum taken from immunodeficiency patients in ICU. (Ethical approval code: 13820507972007)(2). To detect A. baumannii isolates, biochemical experiments and polymerase chain reaction with primers specific for the bla OXA-51 gene were performed (Table 1).
Table 1. Primers sequences and the annealing temperatures used in this study
| Target Genes | Primers sequences (5-3) | Annealing Temperature (ºc) |
DNA amplicon Size (bp) |
Reference |
| OXA-51 | TAATGCTTTGATCGGCCTTG TGGATTGCACTTCATCTTGG |
60 | 324 | (2) |
| aph(2'')-Id | AATCGGTAGAAGCCCAA GCACCTGCCATTGCTA |
58 | 642 | (10) |
| ant(4′)-Ia | CTGCTAAATCGGTAGAAGC CAGACCAATCAACATGGCACC |
58 | 172 | (10) |
| ant(3'')- I | GAAGTACGCAGAAGAGA ACATGGCAAGCTCTAGGA |
60 | 284 | (10) |
| aac(6′)-Ib | TATCCAGCTAAGCGCGAACT ATTTGCCGACTACCTTGGTC |
57 | 490 | (10) |
| aac(3)-I | CTTCAGGATGGCAAGTTGGT TCATCTCGTTCTCCGCTCAT |
57 | 402 | (10) |
| aph(3′)-I | GCTCACGCAACTGGTCCA GA GGCACGCAAGACCTCAACCT |
57 | 816 | (10) |
| aph(2'')-Ib | CTTGGACGCTGAGATATATGAGCAC GTTTGTAGCAATTCAGAAACACCCTT |
60 | 867 | (10) |
| aph(2'')-Ic | CCACAATGATAATGACTCAGTTCCC CCACAGCTTCCGATAGCAAGAG |
58 | 444 | (10) |
Susceptibility testing of antimicrobial agents
As described in a previous study, sensitivity to antimicrobial drugs was determined by disk diffusion method (2). The antimicrobial disks used are as follows: gentamicin (10µg), ampicillin-sulbactam (10/10), ceftazidime (30µg), imipenem (10µg), tobramycin (10µg), doxycycline (30µg), ciprofloxacin (5µg), co-trimoxazole (1.25/23.75µg), piperacillin (100µg), levofloxacin (5µg), and cefepime (30µg) (MAST, Merseyside, UK). Interpretations were made under CLSI guidelines (13). Multidrug-resistant bacteria are resistant to more than two categories of antibiotics. Acinetobacter baumannii was one such organism. Those isolates of A. baumannii that were resistant to more than one antimicrobial agent in all but two antimicrobial categories were considered extensively drugresistant (XDR) (14).
DNA extraction
QIAGEN DNA Mini Kit was used to isolate genomic DNA from bacterial cells after overnight culture. (QIAGEN Inc., Valencia, CA). At 260 and 260/280 nm, a spectrophotometer (ND-1000, Nano-Drop Technologies, Wilmington, DE) was used to measure the amount of DNA in each sample.
Genes associated with aminoglycoside resistance
We used the PCR method disclosed previously to examine the genes associated with aminoglycoside resistance of aph(2'')-Id, ant(4'')-Ia, ant(3'')-I, aac(6'')-Ib, aac(3)-I, aph(2'')-I, aph(2'')-Ib, and aph(2'')-Ic(8). The sequence of primer pairs (Metabion, Germany) is shown in Table 1. The PCR experiments were conducted with PCR Master Mix (Amplicon, Denmark), containing Taq DNA polymerase, deoxynucleotides, MgCl2, and the proper buffer. It took 25 μl for each reaction, containing 12.5 μl of master mix, one μl each of reverse and forward primers (at 100 nM each), 1µl of DNA sample of 200 ng/µl concentration and nuclease-free water for PCR to reach the final volume. The Gene Atlas 322 system (ASTEC) was used for amplification. PCR steps were as follows: initial denaturation step (94ºC, 5 min), then, 94ºC, 1 min for denaturation (30 cycles), 1-minute annealing (Table 1 shows Tm for each primer pairs), and extension at 72ºC for 1 min, then a final extension at 72ºC for 10 minutes. 1% agarose gel electrophoresis was performed to separate DNAs according to their size. Neutral Red (Sigma Aldrich, Germany) was used for DNA staining, UV transillumination was then used to visualize DNA fragments.
ERIC-PCR
ERIC-PCR was performed according to the protocol described previously (15). The oligonucleotide pairs ERIC-1 5'-ATGTAAGCTCCTGGGGATTCAC-3' and ERIC-2 5'-AAGTAAGTGACTGGGGG-3' were utilized to amplify the ERIC like sequences in A. baumannii DNA. The following PCR program was performed: incipient denaturation (94ºC, 10 min), followed by denaturation at 94ºC for 1 min (30 cycles), 1 min annealing at 52ºC, extension (65ºC, 8 minutes) and 16 min extension at 65ºC. For electrophoresis of amplicons generated by PCR, 20 µl of each amplified sample was loaded in 2% agarose gel. Numerical taxonomy and multivariate analysis were used (NTSYS version 2.1; Exeter Software, New York, NY, USA) to analyze ERIC patterns. The ERIC PCR gels Dendrograms were generated using Dice similarity coefficients, arithmetic averages, 1% optimization, and 1% position tolerance. The most similar isolates were deemed clonally related if their similarities exceeded 96%.
Statistical analysis
The version 17.0 of SPSS (SPSS, Inc., Chicago, IL) was used to analyze the data. The statistical significance of the information was calculated by an X2 test. P <0.05 was regarded as significant.
Isolates: their features
Out of the100 A.baumannii isolates, 26%, 24%, 25%, 15%, 10% were recovered from blood, sputum, secretions collected from thoracic tubes (thoracic catheters), wound swabs, and urine, respectively.
Susceptibility to antimicrobial agents
As described previously (2), 67% and 63% of isolates showed gentamicin and tobramycin resistance, respectively. A. boumannii isolates showing resistance to three antimicrobial agents or more were considered MDR. The isolates were also 32% resistant to all antibiotics tested, and 91% were classified as XDRs. The most common resistance pattern was "ampicillin/sulbactam-ceftazidime-imipenem-gentamicin-tobramycin-doxycycline-ciprofloxacin-levofloxacin-cotrimoxazole-piperacillin-cefepime" with 32% frequency.
Genes involved in aminoglycoside resistance
Each isolate contained one or more aminoglycoside resistance genes is shows the frequency of resistance-related genes (Table2). There were three common aminoglycoside resistance genes: aac(6′)-Ib (79%), ant(3'')-I and aph(2'')-Id (47%). All isolates resistant to aminoglycoside had aac(6′)-Ib gene. The phenotypic patterns of aminoglycoside resistance genes in isolates of A. baumannii are shown in Table 3. 64% of isolates had three or more aminoglycoside resistance-related genes simultaneously. Also, 6% of isolates had 6 aminoglycoside resistance genes whose patterns were "aph(2'')-Ib+ aph(2'')-Ic+ aph(2'')-Id+ ant(4′)-Ia+ ant(3'')-I+ aac(6′)-Ib"(Table3).
Table 2. Frequency of aminoglycoside resistance genes in A. baumannii isolates
| Aminoglycoside resistance genes | No = (%) of isolates |
| aph(2'')-Id |
47 |
| aph(2'')-Ib |
37 |
| aph(2'')-Ic |
39 |
| aac(6′)-Ib |
79 |
| aac(3)-I |
5 |
| ant(3'')-I |
47 |
| ant(4′)-Ia |
38 |
| aph(3′)-I |
9 |
Table 3. Patterns of aminoglycoside resistance genes among A. baumannii isolates
| No. of resistance genes | Patterns of aminoglycoside resistance genes | Percent (%) of A. baumannii | Total (%) | ||
| 1 gene | aac(6′)-Ib aph(2'')-Ib ant(3'')-I aph(2'')-Ic aph(2'')-Id ant(4′)-Ia |
18 1 6 1 2 1 |
29 |
||
| 2 genes | aph(2'')-Ib+ ant(4′)-Ia aph(2'')-Ib+ ant(3'')-I aph(2'')-Ib+ aac(6′)-Ib ant(4′)-Ia+ aac(3)-I aac(6′)-Ib+ ant(3'')-I aph(2'')-Id + ant(4′)-Ia |
1 1 1 1 2 1 |
7 |
||
| 3 genes | aph(2'')-Ib+ aph(2'')-Ic+ aac(6′)-Ib aph(2'')-Ib+ aph(2'')-Ic+ ant(3'')-I aph(2'')-Ic+ aac(6′)-Ib+ ant(3'')-I ant(3'')-I+ aac(6′)-Ib+ aph(3′)-I aph(2'')-Ib+ aph(2'')-Id+ aac(6′)-Ib aph(2'')-Ib+ ant(3'')-I+ aac(6′)-Ib aph(2'')-Id + ant(4′)-Ia+ aac(6′)-Ib aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib aph(2'')-Id+ ant(3'')-I+ aac(6′)-Ib aph(2'')-Ib + aph(2'')-Ic+ aph(2'')-Id |
2 2 3 2 1 1 4 2 2 4 1 |
24 |
||
| 4 genes | aph(2'')-Ib + aac(6′)-Ib+ ant(3'')-I+ aph(3′)-I aph(2'')-Ib+ aph(2'')-Ic+ aph(2'')-Id+ ant(3'')-I aph(2'')-Ib+ aph(2'')-Ic+ ant(3'')-I+ aac(6′)-Ib ant(4′)-Ia+ aac(6′)-Ib+ ant(3'')-I+ aac(3)-I aph(2'')-Id+ ant(4′)-Ia+ ant(3'')-I+ aac(3)-I aph(2'')-Ib+ aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib+ aph(3′)-I aph(2'')-Id+ ant(4′)-Ia + aac(6′)-Ib+ ant(3'')-I aph(2'')-Ib + aph(2'')-Id+ ant(4′)-Ia + aac(6′)-Ib |
2 1 2 2 2 2 1 5 1 |
18 |
||
| 5 genes | aph(2'')-Ib+ aph(2'')-Id+ ant(4′)-Ia + aac(6′)-Ib+ ant(3'')-I aph(2'')-Ib + aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib+ aph(3′)-I aph(2'')-Ib + aph(2'')-Ic+ aac(6′)-Ib+ ant(3'')-I+ ant(4′)-Ia aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib+ ant(3'')-I+ ant(4′)-Ia aph(2'')-Ib +aph(2'')-Ic+aph(2'')-Id+aac(6′)-Ib+ ant(4′)-Ia aph(2'')-Ic+ aph(2'')-Id+ aac(6′)-Ib+ ant(4′)-Ia + aph(3′)-I |
2 2 4 2 4 2 |
16 |
||
| 6 genes | aph(2'')-Ib+ aph(2'')-Ic+ aph(2'')-Id+ ant(4′)-Ia+ ant(3'')-I+ aac(6′)-Ib | 6 | 6 |
||
ERIC-PCR
ERIC-PCR produced multiple bands which were different in size ranging from 100to 3000 bp. The patterns are sorted into six types (A-F) and 20 subtypes. Band patterns illustrated that 16 out of 20 subtypes were distinctive ERIC-PCR clusters and four singleton isolates. 13% and 10% of sets were A3/A4 and B3, respectively, the most common among A. baumannii isolates (Table4, Figure 1).
Table 4. ERIC-PCR patterns among A. baumannii isolates
| ERIC Types |
Subtypes | Size of ERIC-PCR products (bp) | Total isolates number (%) |
| A (2 bands) (40%) |
A1 A2 A3 A4 A5 |
550, 1000 100, 800 150, 900 900, 1500 900, 2500 |
3 8 13 13 3 |
| B (3 bands) (19%) |
B1 B2 B3 B4 |
900, 1200, 1500 200, 850, 1500 500, 850, 1000 400, 550, 800 |
3 5 10 1 |
| C (4 bands) (20%) |
C1 C2 C3 C4 C5 |
300, 700, 1000, 1500 700, 850, 1500, 2500 600, 850, 950, 1500 150, 350, 850, 1400 150, 650, 1000, 1500 |
3 5 3 5 4 |
| D (5 bands) (15%) |
D1 D2 D3 |
600, 700, 900, 1200, 1500 300, 600, 900, 1000, 1500 150, 550, 900, 1400, 2000 |
8 6 1 |
| E (6 bands) (5%) |
E1 E2 |
100, 400, 700, 1000, 1500, 3000 300, 550, 700, 850, 1000, 1500 |
1 4 |
| F (7 bands) (1%) |
F1 | 150, 600, 800, 950, 1500, 2000, 2500 |
1 |
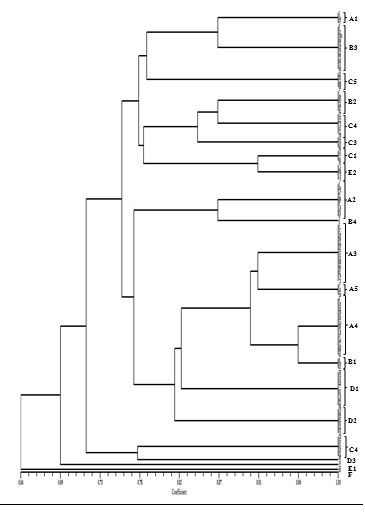
Figure 1. Dendrogram of enterobacterial repetitive intergenic consensus–polymerase chain reaction (ERIC–PCR) of A. baumannii isolates. Scale represents percentages of similarity.
Discussion
A. baumannii is known as the leading cause of nosocomial infections in hospitals as it can acquire a wide spectrum of antimicrobial resistance (2, 3). Limited treatment options have made MDR A. baumannii hard to cure in many environments (10, 16-18). Colistin and tigecycline are the ultimate treating choices (2). This work revealed that 100% isolates of A. baumannii were MDR that were in line with publications from Iran .Studies conducted in Iran (10, 19, 20). In concordance with these results, Safari et al., (2017) found 98% of A. baumannii isolates to be XDR (20). Resistance to Tobramycin and Gentamicin was seen in 67% and 63% of isolates, respectively. The high frequency of aminoglycoside-resistant A. baumannii in our study and previous reports may be due to acquiring AMEs (21). It has been reported that A. baumannii has a wide range of aminoglycoside resistance-related genes. Various risk factors are indicated to be related to the emergence of aminoglycoside-resistant A. baumannii, such as widespread use of antimicrobial agents and inappropriate prescription of aminoglycosides (10). Moreover, the aac(6′)-Ib, aph(2'')-Id, and ant(3'')-I genes showed high prevalence than other genes in this study. Furthermore, the co-existing aac(6′)-Ib and ant(3'')-I was discovered in 33% of isolates. Isolates of A. baumannii in China showed that aac(6′)-Ib and ant(3'')-I are high frequency genes associated with resistance to aminoglycosides found, supporting this study's results (22). According to Haldorsen et al., study (2014) aac(6′)-Ib enzyme could alter amikacin, even in isolates that were phenotypically susceptible to amikacin- of E. coli and Klebsiella spp. (23).
A study conducted by Helmy and Kashef (2017) illustrated aac(6′)-Ib gene in 84.4% of aminoglycoside-resistant Enterobacteriaceae (21). A gene cassette, aac(6′)-Ib, is mainly associated with insertion sequences like IS26 and truncated integrons, class 1 integrons, which probably explains its frequency among A. baumannii isolates (24). Our findings are in line with previous pieces of literature which reported the aac(6′)-Ib and ant (3'')-I as high frequency genes in isolates of A. baumannii (9, 10, 22). In contrast, aac(3)-I and aph(3′)-I genes were less common in this study (8, 22). Similar to this study's results, Xiao et al., (2014) also reported the lower frequency of aac(3)-I and aph(3′)-I genes (16).
The molecular methods for typing A. boumannii are the most critical procedures to determine the association between genetics and epidemiology. ERIC-PCR is a reliable and rapid technique for evaluating genetic differences or clonal relatedness of A. baumannii isolates (11, 12). The ERIC-PCR displayed 2-7 bands (100-3000 bp) based on our findings. The patterns produced by ERIC-PCR were sorted into six types (A-F) and 20 subtypes. They have implied 16 distinctive clusters and four singleton isolates. The most frequent clusters among A. baumannii isolates were A3/A4 (13%) and B3 (10%). In a study conducted by Viana et al., (2011), typing by ERIC–PCR showed a high range of clonal diversity (38 different clones), indicating that A. baumannii spreading depends on multiple factors not entirely attributed to a predominant clone (11). In agreement with our findings, Codjoe et al., (2019) have shown that ERIC-PCR typing of Gram-negative bacilli resistant to carbapenem generates some bands (1 to 8) with different sizes (50-800 bp). Among the 111 carbapenem-resistant (CR) isolates studied, 93 complex dissimilarities were detected, while 18 similar band patterns were observed in pairs (12). Additional studies indicated that A. baumannii showed genetic diversity and heterogeneity. Falah et al., (2019), by identifying 14 different ERIC patterns (ERIC-types) including 11 regular types and three unique types, reported clonal diversity in 80 A. baumannii isolates (25). This study was contrary to Ece et al., (2015) who reported a clonal relationship between strains of A. baumannii collected from a tertiary care center in Turkey. Their results revealed that many A. baumannii clusters in the ICU were the main clusters (26).
Conclusion
Our work indicated the abundance of aminoglycoside-resistant A. baumannii in patients in ICU, as well as the high frequency of aminoglycoside- modifying enzymes: aac(6′)-Ib, ant(3'')-I and aph(2'')-Id. ERIC-PCR has also illustrated an increased level of diversity in A. baumannii isolates. Thus, it can be a convenient method for investigating A. baumanni to determine its isolates' genetic differences or clonal relatedness in clinical environments.
Acknowledgements
This work as PhD thesis in Microbiology was supported by Department of Microbiology, Zanjan Branch, Islamic Azad University, Zanjan, Iran.
Conflicts of Interest
The authors declare that they have no competing interests.
Received: 2021/07/25 | Accepted: 2022/04/6 | Published: 2022/10/10
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






