BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://journal.zums.ac.ir/article-1-5955-en.html
2- Food Microbiology Research Center, Tehran University of Medical Sciences, Tehran, Iran
3- Division of Food Microbiology, Dept. of Pathobiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran ,
✅ Our finding showed that contamination of packed dehydrated vegetables was higher than unpacked; this might be due to drying as well as packaging process. We found that these isolates were negative for enterotoxin.
Clostridium perfringens ranks among the most widespread bacteria, with an ubiquitous environmental distribution in soil, sewage, food, feces, and the normal intestinal flora of humans and animals (1, 2). According to the Center for Disease Control and Prevention (CDC) estimation, more than one million people are infected by C. perfringens each year (3). Food poisoning can be caused by C. perfringens enterotoxin (CPE) produced by C. perfringens spores in the small intestine, which can germinate in foods such as meat and poultry. Main symptoms of the disease are nausea, abdominal pain, and diarrhea. The disease is usually mild and self-limiting in healthy individuals, with symptoms resolving within 24 hours (4,5). The CPE is encoded by its gene located on the chromosome or plasmid of the bacterium (6-8). Expression of the CPE is an important determinant of C. perfringens causing food poisoning (9). Available diagnostic tests for the toxin detection are immunoenzymatic assays including latex agglutination, immunodiffusion, immunoelectrophoresis , ELISA, and Western blot. All these tests depend on high quantity of the enterotoxin in samples. Some strains may carry a silent CPE gene, resulting in missed identification of toxigenic strains. One of the reliable and useful methods for the detection of C. perfringens toxins is polymerase chain reaction (PCR). This method does not depend on enterotoxin concentration (10). Many of C. perfringens food poisoning events occur due to the consumption of contaminated meat and poultry products (11). Detection of toxigenic C. perfringens is important because spore forming bacteria are Stable in the environment. The dehydrated vegetables are routinely used in Iran in various foods, including main dishes and herbal medicines. The aim of this study was to isolate and identify C. perfringens in dehydrated vegetables in Iran.
Sampling
A total of 140 samples of dehydrated green leafy vegetables, including dill, parsley, coriander, tarragon, mint, and mixed vegetables (crumb and soup) were collected from different areas of Tehran, Iran. Samples were collected as unpacked dehydrated vegetables (n=70) and packed (n=70). The study was approved by the Ethics Committee of Tehran University of Medical Sciences (code=IR.TUMS.SPH.REC.1394.775).
Culture and Isolation
To evaluate the samples, 10 g of dehydrated vegetables was diluted in 90 mL of sterile 0.1% peptone water, then 10 mL of this suspension was added to 10 mL of thioglycollate medium. Two thioglycollate tubes were prepared from each sample, one tube was incubated at 75ᵒC for 30 min for the detection of C. perfringens spores and the second tube was incubated at 37°C in an anaerobic jar with Anaerocult A for 48 h. After enrichment step, the supernatant was discarded, and two drops of the pellets were streaked on sulfite polymyxin sulfadiazine (SPS) agar plates and incubated at 37ᵒ C for 24-48 h in an anaerobic jar. Black colonies on SPS agar were subjected to Gram staining and biochemical tests.
Biochemical Tests
Black colonies were assessed for nitrate reduction, gelatinase, production of double zone, motility test, lecithinase production in egg yolk agar, and acid production by lactose and glucose fermentation. The isolates confirmed as C. perfringens were stored at -20ᵒC.
PCR
For genomic DNA extraction, 5-6 colonies grown on SPS agar were inoculated in 200 μl of sterile double distilled water and boiled at 100˚C for 5-10 min. Following the centrifugation at 12,000 g for 3 min, the supernatant was used as the template DNA for PCR assay (12, 13). The specific primers were used to amplify 324 and 233 base pair (bp) of C. perfringens alpha-toxin (CPA) and CPE genes, respectively (Table1). Reference strain of C. perfringens strain CIP 106157 was used as a positive control for detection of CPA and CPE genes. The PCR was performed in a total volume of 20 μL containing 10 μL of master mix (Pishgam, Iran), 10 ng of DNA, 8.5 μL of H2O, and 0.25 mM of each primer (Bioneer, Seoul, South Korea). Thermo cycler (Peqlab, Germany) was set with the following conditions: initial denaturation at 95°C for 5 min and 30 cycles including denaturation at 94°C for 1 min, annealing at 55°C for 2 min, extension at 72°C for 3 min, and final extension at 72°C for 4 min (14). Electrophoresis was performed in 1% agarose gels (Invitrogen, USA) and gel was stained with 0.5 μg ml-1 ethidium bromide (Sigma, USA).
Nucleotide Accession Number
The partial sequence of CPA gene was deposited in GenBank under the accession number of KU166870.
Table 1. Specific primer sequences targeting cpa and cpe
| Reference | Amplicon size (bp) | Sequence(5′-3′) | Gene |
| (9) | 324 | GCTAATGTTACTGCCGTTGA CC TCTGATACATCGTGTAA |
Cpa |
| (9) | 233 | GGAGATGGTTGGATATTAGG GGACCAGCAG TTGTAGATA |
Cpe |
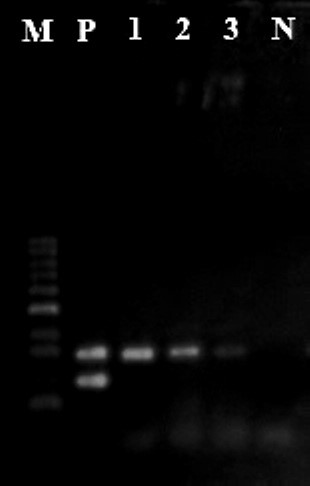
Out of 140 samples, 13 (9.3%) were identified as contaminated with C. perfringens. Among the contaminated samples, 9 (12.8%) and 4 (5.7%) samples were from packed and unpacked samples, respectively. Based on dried vegetables breeds showed no contaminations in soap vegetables, while 1 (0.7%) parsley, 2 (1.4%) coriander, 2 (1.4%) mint, 2 (1.4%) tarragon, 3 (2.2%) dill, and 3 (2.1%) vegetable crumble samples were contaminated with C. perfringens. Using duplex PCR, all isolates were positive for CPA gene while all were negative for CPE gene (Figure 1). This finding indicated that the rate of C. perfringens was 12.8% on the packed dehydrated vegetables and 5.7% on unpacked vegetables.
Figure1. Duplex PCR for cpa and cpe genes. M: marker 100bp, P: CIP106157 positive control for cpa and cpe genes. N: negative control. Line
Discussion
Despite the low frequency of CPE positive and C. perfringens in foods especially in meat, poultry, and dried vegetables, these products can be sources of toxins. Various reports showed that the rate of C. perfringens-associated food poisoning varies around the world (11,16, 17). Unfortunately, there is no comprehensive data showing the frequency of C. perfringens in food poisoning in Iran. This could be due to various reasons including difficulty in registration of patient data or short period of symptoms (24 h). In this study, we determined that 9.3% of dehydrated vegetables were contaminated with C. perfringens. This contamination might be due to the presence of spores in dehydrated samples. The spores are resistant to desiccation, heating, and other conditions. In 2009, Sagoo et al. reported that C. perfringens contamination in dried vegetables and spices is 0.4%. In some other studies conducted in the UK (1975, 1985, 1986), the isolation rate of C. perfringens isolation from dried vegetables was 0 to 7.6% (15).
The frequency of C. perfringens isolated from packed vegetables was more than unpacked ones. This finding may be related to inappropriate processing during packaging or lack of hygiene consideration in factory, transportation, and packaging. The isolates were investigated for CPA and CPE genes. Duplex PCR was applied for 13 confirmed isolates, in which all samples were positive for CPA gene and negative for CPE gene. As far as the researchers investigated, no study has been carried out on dehydrated vegetables in Iran yet. But some studies have been carried out in Iran on isolation of C. perfringens from poultry meats. Poursoltani et al. (in 2014) detected six isolates from 180 packed poultry samples. They used multiplex PCR for C. perfringens subtyping and single PCR for netB and tpel genes in poultry with necrotic enteritis symptoms (18). In Iran, Zandi et al. (in 2014) showed that 100% ostrich dung samples were positive for CPA gene. The multiplex PCR method was used that have not been reported in Iran (19). The difference in frequency of contamination may be due to different contamination sources. Erol et al. (2008) used multiplex PCR on ostrich meat, and showed that 12.2% (22/180) isolates were positive for CPA gene but negative for CPE gene (12).
Conclusion
Our results showed that 13 (9.3%) samples were contaminated with C. perfringens. However, the bacteria were negative for CPE. The higher contamination rates in packed vegetables compared to unpacked ones might indicate the lack of suitable hygienic considerations on drying and packaging processes.
Acknowledgements
This paper is part of a research project approved by the Food Microbiology Research Center, Tehran University of Medical Sciences and Health Services Contract No. 28439. The authors would like to thank Dr. Zahraei Salehi from Tehran Veterinary University for providing us a C. perfringens strain as control.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Funding and support
This research resulted from an independent research without receiving any financial support.
Conflicts of Interest
Authors declared no conflict of interests.
Received: 2020/03/25 | Accepted: 2020/07/16 | Published: 2020/12/4
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |