BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://journal.zums.ac.ir/article-1-6130-en.html


 , Ahmad Kazemi2
, Ahmad Kazemi2 

 , Bahram Chahardouli1
, Bahram Chahardouli1 

 , Saeed Mohammadi1
, Saeed Mohammadi1 

 , Mohsen Nikbakht1
, Mohsen Nikbakht1 

 , Nasrin Alizadeh1
, Nasrin Alizadeh1 

 , Asadollah Mousavi1
, Asadollah Mousavi1 

 , Kamran Alimoghaddam1
, Kamran Alimoghaddam1 

 , Majid Teremmahi Ardestani *3
, Majid Teremmahi Ardestani *3 


2- Dept. of Hematology, School of Allied Medical Sciences, Iran University of Medical Sciences, Tehran, Iran.
3- Dept. of Medical Technology, School of Allied Medical Sciences, Hormozgan University of Medical Sciences, Bandar Abbas, Iran ,
✅ The results of the current study do not support WT1 mutation, SNP rs16754, or WT1 overexpression at diagnosis, as they were found to be poor prognostic markers in AML patients.
Acute myeloid leukemia (AML) is a genetically and epigenetically heterogeneous disease with acquired mutations (1). Somatic mutations, such as Wilms tumor 1 (WT1), FMS-like tyrosine kinase-3 internal tandem duplication (FLT3-ITD), nucleophosmin (NPM1), and CCAAT/enhancer-binding protein alpha (CEBPA), play important roles in the pathogenesis of AML. WT1 is located at chromosome 11p13, and it has 10 exons, of which exons 7-10 make the zinc finger domain of the WT1 protein with DNA-binding activity (2). WT1 is primarily considered as a tumor suppressor gene and responsible for childhood Wilms tumors. Several studies have shown the oncogenic properties of WT1 in some tumors.
WT1 with anti-apoptotic activity takes part in the expansion of myeloid cells, and its expression is inversely related to cell differentiation. The majority of patients with AML have high WT1 expression levels. In the French-American-British (FAB) AML subgroups, the highest and lowest levels of WT1 expression were seen in M3 and M5, respectively (3). Some studies have shown a prognostic impact of WT1 expression at diagnosis, such that the overexpression of WT1 has been linked to a failure to achieve complete remission (CR) and low overall survival (OS) rate. However, other studies have not confirmed these results (4, 5).
Somatic mutations in WT1 occur in approximately 6%-15% of de novo AML. The majority of mutations occur in exons 7-9 and result in loss of function and expression of a truncated protein lacking the zinc finger domain. WT1 mutations are associated with younger age and the existence of FLT3-ITD and CEBPA mutations. The exact role of WT1 mutations in the pathogenesis of AML is rather controversial. Recent studies have shown that WT1 plays an important role in epigenetic regulation. WT1 (in combination with TET2 and IDH1/2) contributes to the demethylation process. Hence, WT1 mutations can disrupt the demethylation process (6-8).
Single-nucleotide polymorphism (SNP) rs16754 (A to G substitution at nucleotide position 1297) is located in exon 7 of the WT1 gene and apparently confers favorable clinical outcomes in adult AML patients. However, some studies have failed to confirm such an association (9).
Owing to the cited controversy among current reports, in the present study, we assessed the frequency of WT1 mutations, SNP rs16754, WT1 expression levels, and their prognostic impact on patients with AML in an Iranian population.
A total of 130 de novo AML patients (81 males and 49 females) with a median age of 42 years (range=15-79 years), who were referred to the Hematology-Oncology and Stem Cell Transplantation Research Center of Shariati Hospital, were recruited for our study. Informed written consent was obtained from all patients in line with the ethical guidelines of Iran University of Medical Sciences, which approved this study.
Patients were diagnosed based on the existence of blast cells ≥ 20% in bone marrow (BM) smears, and they were categorized into subgroups according to the FAB classification. Immunophenotyping was performed in all cases. Patients with AML-M3 were excluded.
Treatment Regimens
All patients were initially treated with induction chemotherapy (100 mg/m2/d cytarabine every 24 hours [days 1, 3, 5, and 7] and 60 mg/m2 daunorubicin for three days). In order to achieve CR, patients received one or two courses of induction chemotherapy. The definition of CR was defined as ≤ 5% blast in BM, recovery of neutrophils and platelets (PLTs), and absence of extramedullary diseases. In consolidation therapy, allogeneic hematopoietic stem cell transplantation was considered for eligible patients, while the rest were treated with cytarabine (100 mg/m2/day every 24 hours for five days) and daunorubicin (60 mg/m2 for two days). For patients who were refractory or relapsed salvage therapy was administered.
DNA and RNA Extraction
Genomic DNA was extracted by a salting-out procedure. Total RNA was isolated using the Trizol reagent (Qiazol, Qiagen, USA). All protocols were performed according to the manufacturer’s instructions. Further, cDNA synthesis from 1 μg of RNA was performed in a 20 μL reaction using random hexamers and oligo (dT) according to the manufacturer’s instructions using a cDNA synthesis kit (Takara Bio Inc., Otsu, JAPAN).
Real-Time Quantitative Polymerase Chain Reaction of WT1
WT1 expression levels were determined by quantitative real-time polymerase chain reaction (PCR) using the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). A WT1 mRNA assay kit (Qiagen, USA) was used to detect WT1 gene expression. The ABL housekeeping gene was used for normalization. WT1 levels were expressed as copies of WT1/ABL× 104, and a copy number of ≥ 250 was considered as overexpression.
Mutation Analysis
WT1 exons 7 and 9 were amplified using specific primers. PCR conditions were as follows: one cycle of 95°C for three minutes; 35 cycles of 94°C for 30 seconds, 60°C for 20 seconds, and 72°C for 30 seconds; and a final extension of 72°C for seven minutes. The PCR products were analyzed by electrophoresis on a 2.0% agarose gel, and the products were then directly sequenced.
Details of the detection of NPM1, FLT3-ITD, and CEBPA mutations were described previously (10, 11).
Statistical Analysis
The x2 (chi-square) test was used to analyze categorical variables. Continuous variables (age, as well as white blood cell [WBC] and PLT counts) were analyzed using the Mann-Whitney test. The Kaplan-Meier method was used to calculate OS and disease-free survival (DFS). OS was defined as the time from the date of initial diagnosis until death from any cause. DFS was defined as the duration between the date of CR until the date of relapse or death. The log-rank test was used to compare the differences in OS and DFS between different groups.
Frequency and Types of WT1 Patients
Overall, seven mutations were identified in seven of 130 patients (5.4%). All mutations were located in exon 7, and no mutations were found in exon 9. All mutations were of the insertion type and ranged from 1 to 7 bp (Table 1). All WT1 insertion mutations resulted in frameshift mutations; these mutations caused a truncated protein, which altered its ability to bind to DNA. Laboratory data, other molecular aberrations, and clinical characteristics of patients with mutated and unmutated WT1 genes are summarized in Table 2.
No significant differences were found in terms of gender, age, WBC, hemoglobin (Hb), PLT counts, or FAB classification between patients with and without WT1 mutations. Among other molecular aberrations, no correlation was found between WT1 mutations, NPM1 mutations (P=0.360), or CEBPA mutations (P=0.170). However, a correlation was found with FLT3-ITD (P=0.010).
Table 1. Characteristics of AML patients with mutated WT1
| WT1mut7 | Age/sex | FAB type | DNA change | Protein change | Other molecular aberrations | Outcome |
| 1 | 46/F | M4 | 1328-1334dup | S315Vfsx4 | FLT3-ITD | Relapse/Death |
| 2 | 23/F | M1 | 1333-334insCGGT | A314Gfsx4 | FLT3-ITD/NPM1mut | Live |
| 3 | 17/M | M1 | 1327-1331dup | S313Yfs | None | Relapse/Death |
| 4 | 27/M | M2 | 1325-1326insGACTCTT | V311Ffsx6 | FLT3-ITD/CEBPAmut | Relapse/Death |
| 5 | 40/M | M5 | 1330-1331insG | S313Vfsx4 | FLT3-ITD | Relapse/Death |
| 6 | 52/M | M1 | 1334dup | A314Gfsx2 | FLT3-ITD | Live |
| 7 | 48 /F | M1 | 1325-326insGACTCTT | V311Ffsx6 | NPM1mut | Relapse/Death |
The nucleotide numbering is according to the GenBank accession number NM_024426 (transcript variant D). The numbering of amino acid residues is according to SWISS-PROT protein sequence number P19544.
Table 2. Correlation of WT1 mutations and overexpression with clinical characteristics in AML patients
| Clinical variants | Total (n=130) |
WT-1 Mutation (n=7) |
P-value | WT-1 Overexpression (n=108) |
P-value |
|---|---|---|---|---|---|
| Gender: Male Female |
81(62.3) 49(37.7) |
4(57.1) 3(42.9) |
0.6 |
67(62.03) 41(37.97) |
0.545 |
| Age: ≤60 >60 |
121(87.7%) 9 (5.4%) |
7(100) 0 |
0.24 |
100(92.6) 8(7.4) |
0.610 |
| FAB subtypes: | 0.237 | ||||
| M0 | 2(1.5%) | 0 | 0.093 | 2(1.8) | |
| M1 | 19(14.6%) | 4(57.1) | 17(15.7) | ||
| M2 | 47(36.2%) | 1(14.3) | 39(36.2) | ||
| M4 | 45(34.6%) | 1(14.3) | 40(37.05) | ||
| M5 | 12(9.2%) | 1(14.3) | 6(5.55) | ||
| M6 | 4(3.1%) | 0 | 4(3.7) | ||
| M7 | 1(0.8%) | 0 | 0 | ||
| WBC: Median Range |
27408.9 150-294000 |
26235.7 150-91200 |
0.587 |
31046.8 150-294000 |
0.034 |
| Hb: Median Range |
8.8 4.9-14.2 |
8.8 5-14 |
0.778 |
8.73 4.9-14.2 |
0.62 |
| Plt : Median Range |
65681 2000-799000 |
34500 8000-60000 |
0.375 |
64592 2000-799000 |
0.45 |
| Molecular aberrations: | |||||
| NPM1 mutation | 23(17.7%) | 2 (28.5) | 0.360 | 21(19.4) | 0.007 |
| FLT3-ITD | 32(24.6%) | 5(71.4) | 0.10 | 28(25.9) | 0.93 |
| CEBPA mutation | 13(10%) | 1(14.28) | 0.170 | 9(8.3) | 0.175 |
Abbreviations: AML, acute myeloid leukemia; WT1, Wilms tumor 1; FAB, French-American-British; ITD, internal tandem duplication; WBC, white blood cell; PLT, platelet.
Clinical Outcome and Prognostic Impact of WT1 Mutations
According to the standard protocol for induction therapy, of the 130 patients with AML, 94 (72.3%) achieved CR, 23 (17.7%) were resistant, and 13 (10%) died.
In achieving CR, no significant difference was found between patients with and without WT1 mutations (P=0.664). Among other molecular aberrations, patients with and without NPM1 mutations (P=0.45) and patients with and without CEBPA mutations (P=0.263) were similar in their achievement of CR. However, in relation to FLT3-ITD, a statistically significant difference in achieving CR was found between patients with and without FLT3-ITD status (P=0.012).
The five-year OS and DFS rates were calculated between patients with and without WT1 mutations. No significant differences were found in terms of OS (P=0.98) or DFS (P=0.97) between the two groups (Figure 1).
Analysis of WT1 SNP rs16754 in AML Patients
SNP rs16754 in 130 adult AML patients was analyzed. Of the 130 patients, 104 (80%) had genotype AA, 22 (16.9%) had genotype AG, and four (3.1%) patients carried genotype GG. No significant difference was found in terms of gender, age, WBC, Hb, and PLT counts among the various genotypes of SNP rs16754.
No correlation was found between SNP rs16754 and other molecular aberrations. In achieving a first CR after induction therapy, no significant difference was observed among the various genotypes of SNP rs16754 (P=0.490).
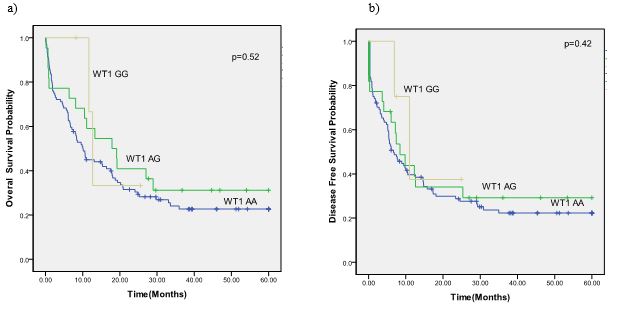
In the next step, OS and DFS in AML patients were compared between the genotypes of SNP rs16754. No statistically significant difference was observed in OS (P=0.52) and DFS (P=0.42) (Figure 2).

Figure 1. The survival curve of AML patients according to the mutational status of WT1. a) DFS in overall patients. b) OS in overall patients
Abbreviations: WT1, Wilms tumor 1; AML, acute myeloid leukemia; DFS, disease-free survival; OS, overall survival.
Figure 2. The survival curve of AML patients according to the SNP rs16754 genotype status. a) DFS in overall patients. b) OS in overall patients
Abbreviations: WT1, Wilms tumor 1; AML, acute myeloid leukemia; SNP, single-nucleotide polymorphism; DFS, disease-free survival; OS, overall survival.
WT1 Expression in AML Patients
The frequency of WT1 overexpression at the time of diagnosis in the current study was 83.1%. The clinical characteristics, laboratory data, and analyses of other molecular aberrations are summarized in Table 1. No significant differences were found between WT1 expression levels and clinical characteristics, except in WBC count (P=0.034). The median expressions of WT1 in relation to WT1 mutations, various genotypes of SNP rs16754, and other molecular aberrations were also assessed.
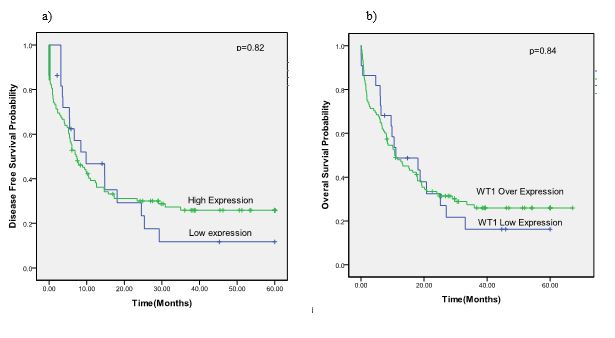
The median expressions of WT1 in AML patients with and without WT1 mutations were compared; WT1 overexpression was seen more often in patients with wild-type WT1, though this difference was not statistically significant (P=0.6). The median WT1 expression level was higher in the genotype AA of SNP rs16754 than in other genotypes, but, again, the difference was not statistically significant (P=0.262). Regarding molecular aberrations, only NPM1 mutations (P=0.007) were associated with WT1 overexpression. No correlation was found between WT1 expression levels and initial CR achievement after induction therapy (P=0.46). Furthermore, no statistically significant difference was found in OS (P=0.84) or DFS (P=0.82) regarding WT1 expression levels (Figure 3).
Figure 3. The survival curve of AML patients according to the WT1 expression level status. a) DFS in overall patients. b) OS in overall patients
Abbreviations: WT1, Wilms tumor 1; AML, acute myeloid leukemia; DFS, disease-free survival; OS, overall survival.
Discussion
Understanding the prognostic impact of molecular aberrations on AML is essential to assessing risk stratification. WT1 gene mutation and expression are among the molecular markers that have been extensively considered in recent years.
In the present study, the incidence of WT1 mutations (5.4%) was lower than in some other studies, including one previous study that was also conducted on an Iranian population (12.5%) (12). Similar to Mittelmann et al., all mutations were of the insertion type and located in exon 7, and no mutations were found in exon 9 (13). Congruent with other studies, no significant difference was found between WT1 mutations and clinical characteristics, such as gender, age, Hb, WBC, and PLT counts (8).
Hou et al. showed that there was no correlation between WT1 mutations and NPM1, FLT3-ITD, and CEBPA mutations (6). In other research, Toogeh et al. failed to find any association between WT1 mutations and FLT3-ITD mutations (12). In this study, neither NPM1 nor CEBPA mutations were correlated with WT1 mutations, whereas a correlation was observed between WT1 mutations and FLT3-ITD positive status (P=0.010).
In contrast to our results, Tien et al. revealed that patients with co-occurrence of WT1 mutations and CEBPA double mutations had a poor prognosis and that BM transplantation may be considered as first-line therapy in CR1 (14).
To assess the prognostic impact of WT1 mutations on AML patients, patients were followed for five years, and the OS and DFS terms were analyzed. Achieving CR and clinical outcomes were compared between the two groups of patients, and no significant difference was found.
In contrast to our results, a series of studies have shown that patients with mutations have lower CR or higher relapse rates than patients without WT1 mutations. Niktoreh et al. declared that WT1 mutations combined with FLT3-ITD show reduced 3y-EFS and OS when compared with patients with either WT1 or FIT3-ITD and patients with neither of these mutations (15). Tanwar et al. evaluated WT1 mutations (exons 7 and 9) in 100 patients with AML; 17 mutations (exon 7=10; exon 9=7) were found. They stated that more patients were needed to confirm the link between WT1 mutations and clinical outcomes (16).
Consistent with the current results, Gaidzik et al. could not show any impact of WT1 mutations on the outcomes of patients with AML (8). The discrepancies between studies may be due to unknown cooperating genetic aberrations or different post-remission consolidation therapies.
Several studies have been conducted on the prognostic impact of SNP rs16754 on adult AML patients, and the results are inconsistent. Damm et al. reported that AG/GG genotypes independently predicted improved outcomes in adult AML patients (17). Ho et al. reviewed 790 pediatric AML patients and revealed that SNP rs16754 independently predicted improved OS and DFS (18). Sheng Lou et al. and Petit et al. also found that the clinical outcomes of adult AML patients with genotype GG are better than those with two other genotypes (5, 19). No prognostic impact of SNP rs16754 on the clinical outcomes of AML patients was found in the current study. Some previous studies were unable to confirm that SNP rs16754 predicts OS and DFS (18, 20).
The frequency of WT1 overexpression in the current study population was 83.1%, which is in agreement with previous research (21). In some studies, WT1 overexpression at diagnosis has been described as an adverse predictor of clinical outcomes. Lyu et al. reported that WT1 overexpression by itself is an independent and negative predictor of CR, DFS, and OS in cytogenetically normal AML patients (22, 23). Du et al. showed that AML patients with high WT1 levels had a lower overall response rate and two-year OS when compared with patients with a low copy number of WT1 (24). Dongfeng et al. showed that AML patients with high level of WT1 had a lower overall response rate and 2-year OS compared with patients with low copy number of WT1 (25). Mitrovic et al. Showed that increased expression of WT1 gene detected in high proportion of APL patients could be considered as a good indicator for risk stratification to improve treatment and outcome of APL patients (26). Ebtesam et al. believed that WT1 overexpression is an independent negative prognostic marker that could be used to evaluate response to induction chemotherapy and prognosis of AML patients (27). Shimada et al. did not find any correlation between WT1 overexpression and poor outcomes in Japanese pediatric AML patients (28). The current study failed to show a significant difference in CR rate, OS, or DFS in AML patients regarding WT1 expression levels.
The current results are in line with those of Luo and Gaur, who indicated that the median expression of WT1 is relatively higher in AML patients without mutations when compared to AML patients with mutations. However, the difference was not statistically significant. The current results also showed that patients with NPM1 mutations have higher levels of WT1 expression than those without NPM1 mutations. Differently from the current findings, Luo et al. did not find any correlation between WT1 expression levels and other genetic alterations, such as FLT3-ITD, CEBPA, or MLL. Gaur et al. also indicated that AML patients with FLT3-ITD or FLT3-TKD mutations had a high median copy number of WT1 transcripts (5, 29).
Conclusion
The results of the present study indicate that WT1 mutations, WT1 expression levels, and SNP rs16754 cannot be considered as prognostic factors in random adult patients with AML. Owing to the low sample size, low frequency of WT1 mutations, and role of unknown cooperating genetic aberrations, the current results must be interpreted with caution. It is suggested that a large-scale study is conducted to confirm the present results.
Acknowledgements
Thanks to the professors and staffs of Hematology-Oncology & Stem cell transplantation Research center, Tehran University of Medical Sciences.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Funding and support
This work was supported by a grant from Iran university of medical sciences [grant number IR.IUMS.REC.1394.26066]
Conflicts of Interest
Authors declared no conflict of interests.
Received: 2020/07/19 | Accepted: 2020/09/24 | Published: 2020/12/4
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |