BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://journal.zums.ac.ir/article-1-6763-en.html
2- Dept. of Genetics, Faculty of Sciences, Zanjan Branch, Islamic Azad University, Zanjan, Iran
3- Dept. of Genetics, Faculty of Sciences, Zanjan Branch, Islamic Azad University, Zanjan, Iran ,
✅No significant correlation was found between the occurrence of mutant alleles of CXCL12 and CXCR4 polymorphisms and breast cancer. The relapse of the disease after chemotherapy and the age of menopause are associated with the occurrence of polymorphisms. However, further studies are needed to confirm the results.
Breast cancer is an uncommon growth of malignant cells in various breast tissues, such as milk transfer ducts, milk-producing glands, and nonglandular tissues of the breast. It is one of the most commonly diagnosed cancers in women worldwide. Breast cancer is the third most common cancer in the world after lung and gastric cancers (1). In 2008, about 1.38 million new cases of breast cancer were diagnosed, and by 2020, it is expected to reach 1.7 million (2). The associated chemokine receptors and chemokine ligands play an important role in cancer modeling, including tumor growth, survival, migration, angiogenesis, and infiltration of immune cells. The interaction of C-X-C chemokine receptor 4 (CXCR4)/C-X-C motif chemokine ligand 12 (CXCL12) is the most common interaction seen in many malignancies, such as breast, ovarian, and prostate cancers (3).
The CXCL12 gene, also known as stromal cell-derived factor 1 (SDF-1), is located on human chromosome 10 (4, 5). It interferes with tumor pathogenesis by increasing tumor growth, interfering with the migration and adhesion of tumor cells, enhancing angiogenesis, and providing an immunosuppressive microenvironment. The related protein is a ligand for coupled protein receptor G (6). CXCL12 has a chemotactic function in lymphocytes (7). During the fetal period, it leads to the transformation of hematopoietic cells from the liver into the bone marrow and the formation of large blood vessels. It has also been shown that the signaling of CXCL12 regulates the formation and expression of CD20 in B cells (8). CXCL12 is also chemotactic for mesenchymal stem cells; it is formed in the region of inflammatory bone destruction and moderates the suppressor effect of this region on osteoclast genesis (9). In adulthood, CXCL12 plays an important role in angiogenesis by absorbing endothelial progenitor cells (EPCs) from bone marrow by a mechanism dependent on CXCR4 (10). CXCL12 is expressed on stromal cells (fibroblasts and endothelial cells), especially bone marrow, liver, and peritoneum cells. Hypoxia, tissue damage, and inflammation increase the expression of CXCL12 and its receptor (ie, CXCR4) (11, 12).
The CXCL12 (rs1801157) polymorphism (named G801A) is located on the 801st pair of the 3' untranslated terminal region (13). A significant frequency of the CXCL12 rs1801157 polymorphism AA genotype was observed in patients with renal cell carcinoma, in which the overall survival was shorter compared with patients presenting GG and GA genotypes. According to some studies, AA and GA genotypes are more susceptible to distant metastasis development in esophagogastric cancer (14).
CXCR4 (also known as Fusin or CD184) is a protein with 7 membrane regions on the cell surface. CXCR4 is a kind of receptor for CXCL12 (15). The CXCR4 gene is located on 2q22.1 and has a length of 10 807 base pairs (bp). It has 2 exons and 1056 nucleotides in the coding region of the gene. CXCR4 is one of the several chemokine receptors that HIV can use to influence T cell CD4+. CXCR4 is positively regulated during implantation procedures in normal periods and hormone replacement in the endometrium; in the presence of human blastocysts, it leads to the polarization of a part of the CXCR4 receptor, indicating the role of this receptor at the implantation stage in humans. There is a single nucleotide polymorphism (SNP; rs2228014) on codon 138 of CXCR4; this receptor is an interesting tumor marker, especially in the metastatic process (16).
So far, SNPs associated with breast cancer have been identified. In general, the combination of rs2228014 (CXCR4) and rs1801157 (CXCL12) polymorphisms is associated with breast cancer and the survival of patients in a variety of cancers. The expression level of the CXCR4 gene polymorphism is also associated with the invasion of cancer and its prognosis. Studies have shown that the rs1801157 GA/AA polymorphism has a significant relationship with distant metastasis. To our knowledge, no study has been conducted on the association between CXCR4 (rs2228014) and CXCL12 (rs1801157) polymorphisms with metastatic breast cancer and the therapeutic outcome of patients with chemotherapy drugs in the Iranian population. Since CXCR4 and CXCL12 gene polymorphisms can serve as candidate agents in detecting prognostic factors for molecular evaluation and treatment of patients with metastatic breast cancer, we evaluated the prognostic role of these polymorphisms and the therapeutic outcome of patients undergoing chemotherapy with the occurrence of these polymorphisms.
Sampling
In this case-control study, 30 patients aged 25-70 with metastatic breast cancer type IDC under chemotherapy and 30 healthy nonrelatives with no history of breast cancer or any other cancers were studied according to their age. Demographic characteristics (including age, sex, smoking, history of alcohol use, etc), related factors (including consumption of sweets/fat, family history of cancer, history of abdominal surgery, etc), and some important issues (including the type of chemotherapy drugs, their therapeutic response, and the type and degree of their tumor) were collected using questionnaires. This research was approved by the Ethics Committee of Zanjan University of Medical Sciences (code: ZUMS.REC.1395.138). Informed consent was obtained from all participants.
Genotype Determination
Using a specific DNA extraction kit (SinaClon, Iran), genomic DNA was extracted from the peripheral blood of patients and normal individuals. Subsequently, the polymerase chain reaction (PCR) was performed using specific primers of CXCR4 and CXCL12 genes (Table 1) by the tetra-primer amplification refractory mutation system–PCR (T-ARMS–PCR) method. The total reaction (25 μL) contained 100 ng of genomic DNA (1-4 μL), Master Mix 2X Taq Premix (CinnaGen, Iran) at a concentration of 12.5 μL of each primer (1.25 μL), and 6.5 μL of deionized water (Table 1). The reaction condition consisted of preheating at 95 °C for 5 minutes, followed by 30 cycles of denaturation at 95 °C for 30 seconds, annealing at 64.7 and 62.7 °C for CXCR4 and CXCL12 genes for 45 seconds, extension at 72 °C for 30 seconds, and final extension at 72 °C for 5 minutes. Scoring was done by running the PCR products on a 2% agarose gel electrophoresis at 3-5 V/cm for 40 minutes.
Statistical Analysis
Allele and genotype frequency analyses in all subjects were analyzed using SPSS version 23 (SPSS Inc Chicago, I11, USA). The Fisher exact and chi-square tests were also used for statistical analyses. P values less than 0.05 were considered statistically significant.
Table 1. The sequence of primers and the PCR products of CXCL12 (rs1801157) and CXCR4 (rs2228014) polymorphisms
| Temperature of primers (°C) | Product length | Primer sequence | SNP |
| 67.2 |
251 bp | FIP: 5'- TCATCAGTCTGGACCGCTACCTGGCCCTC- 3' |
CXCR4 (rs2228014) C/T |
| 65.9 |
208 bp | RIP: 5'- GCCTCTGACTGTTGGTGGCGTGGCCA- 3' | |
| 67.2 |
404 bp |
FOP: 5'- CTGCACCTGTCAGTGGCCGACCTCCTCT- 3' | |
| 65.7 |
ROP: 5'- CCAGGCAGGATAAGGCCAACCATGATGTGC-3' | ||
| 67.2 |
200 bp | FIP: 5' -ACCCCCTTCTCCATCCACATGGGAGACA- 3' |
CXCL12 (rs1801157) G/A |
| 65.9 | 275 bp | RIP: 5' -GCTGCCCTCCCAGAAGAGGCAGAACC- 3' | |
| 65.8 |
421 bp |
FOP: 5' -TTCCACGGAGCCACTCCTCTGACTCAGG- 3' | |
| 64.3 |
ROP: 5' -GCCATGGAGACAGTCGTGGACACACATG- 3' |
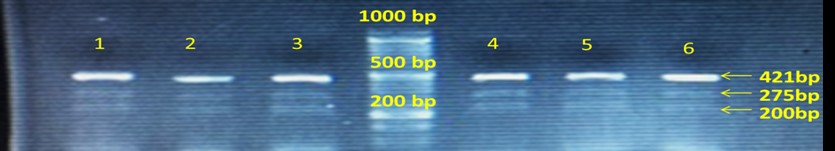
(A1) The CXCL12 gene: 50-bp ladder, the 421-bp band as a specific outer control in all samples. Two bands of 421 and 275 bp are normal homozygous genotype GG, and dual bands at 421 and 200 bp represent the homozygous genotype of mutant AA. All three 421-, 275-, and 200-bp bands show heterozygous genotype AG (lanes 1-6).
(B1) The CXCR4 gene: 50-bp ladder, the 404-bp band as an outer control in all samples. Two bands of 404 and 252 bp are the normal homozygous genotype (lanes 1-9), and double bands at 404 and 208 bp represent the homozygous mutant TT genotype. All three 404-, 252-, and 208-bp bands represent heterozygous genotype CT.
Genotyping Results of CXCR4 and CXCL12 Genes
The average age was 50 years in the patient group (n = 30) and 41.23 years in the control group (n = 30). Based on the statistical results, a significant relationship was found between age and incidence (P = 0.02). The results showed that in the patient group (n = 30), 63.33% of patients were diagnosed with breast cancer before menopause and 36.66% after menopause. Also, the body mass index (BMI) of the patients was between 22%-25% and 33.3%. We found that 73.32% of patients were diagnosed with metastasis 3-6 years after primary chemotherapy, and 63.33% had grade III tumors (Table 2).
Table 3 shows genotype frequencies in CXCR4 and CXCL12 genes. The statistical analysis showed no significant difference between the rs2228014 polymorphism of the CXCR4 gene (P = 0.49) and the rs1801157 polymorphism of the CXCL12 gene (P = 1) in both groups.
The Relationship Between the Allele Frequencies of CXCL12 and CXCR4 Polymorphisms and Breast Cancer
The results of normal and mutant alleles in both groups for the CXCL12 polymorphism (rs1801157) revealed that the frequency of G and A alleles was 65% and 35% in the patient group and 63.33% and 36.66% in the control group. As a result, based on the statistical analysis, no significant correlation was found between the 2 groups in the occurrence of normal and mutant alleles for the CXCL12 polymorphism (rs1801157; P = 0.84 and P < 0.05). The results of normal and mutant alleles in both groups for the CXCR4 polymorphism (rs2228014) showed that the frequency of C and T alleles was 100% and 0% in the patient group and 95% and 5% in the control group. As a result, based on the statistical analysis, no significant correlation was found between the 2 groups in the occurrence of normal and mutant alleles for the CXCR4 polymorphism (rs2228014; P = 0.18 and P < 0.05).
Also, the results of normal and mutant alleles in both groups for rs1801157 and rs2228014 polymorphisms indicated that the number of normal alleles was 16.66% in the patient group and 15% in the control group; there were no significant correlations between the 2 groups in the simultaneous occurrence of normal and mutant alleles for rs1801157 and rs2228014 polymorphisms (P = 0.95 and P < 0.05; Table 2).
Table 2. Genotypic and allelic frequency in XRCC5 and XRCC6 genes
| SNP rs2228014 | Genotype frequency in the patient group (n = 30) |
Genotype abundance in the control group (n = 30) |
P value | |
| Genotypes, n (%) | ||||
| CC | 30 (100) | 28 (93.3) | 0.472 | 0.49 |
| CT | 0 | 1 (3.33) | 1 | |
| TT | 0 | 1 (3.33) | 1 | |
| Allele, n (%) | ||||
| C | 60 (100) | 57 (95) | 0.1898 | |
| T | 0 | 3( 5) | ||
| SNP rs1801157 | Genotype frequency in the patient group (n = 30) |
Genotype abundance in the control group (n = 30) |
P value | |
| Genotypes, n (%) | ||||
| GG | 10 (33.3) | 10 (33.3) | 1 | 1 |
| AG | 1 (3.33) | 2 (6.66) | 1 | |
| AA | 19 (63.3) | 18 (60) | 1 | |
| Allele, n (%) | ||||
| G | 39 (65) | 38 (63.33) | 0.8490 | |
| A | 21 (35) | 22 (36.66) | ||
| The simultaneous occurrence of normal C and G alleles |
20 (33.33) | 18 (30) | 0.9596 | |
| Co-occurrence of mutant A and T alleles |
0 | 0 | ||
The Relationship Between the Genotyping Frequencies of CXCL12 and CXCR4 Polymorphisms and Clinical Characteristics of Patients
According to the literature, various results have been obtained regarding the effect of CXCR4 and CXCL12 polymorphisms on the increased risk of breast and other cancers, as well as regarding the association of the polymorphisms of these 2 genes with distant metastases. In some studies, both have significant effects, and in other studies, only one of these polymorphisms had a significant effect. In the present study, no significant relationship was observed between the incidence of T and A mutants and the increasing risk of the disease.
According to the clinical information of patients, we found a significant relationship between the time of metastasis, the recurrence of the disease after initial chemotherapy, and the occurrence of the CXCL12 gene polymorphism (rs1801157) and CXCR4 gene polymorphism (rs2228014; P = 0.001 and P < 0.05). The results showed that in the patient group (n = 30), 63.33% of patients were diagnosed with breast cancer before menopause and 36.66% after menopause. According to the statistical analysis, the relationship between menopausal age and the incidence of the disease was near the significant level (P = 0.07 and P < 0.05). Thus, affected individuals with late menopause had a small increased risk of developing breast cancer. The reason could be that they were exposed to estrogen for longer periods compared with women who started their periods later or had early menopause. The mean BMI was 25.21 kg/m2 in the patient group, showing no significant correlation between BMI and the incidence of genotypes (P = 0.899 and P < 0.05). In the present study, the association between the age of patients and the genotype of CXCR4 and CXCL12 genes was also studied. The statistical results indicated a significant relationship between age and the occurrence of the rs1801157 polymorphism of the CXCL12 gene (P = 0.02 and P < 0.05); therefore, age is the most significant risk factor for developing breast cancer. Most breast cancers occur in women over the age of 50. In contrast, there was no significant relationship between age and the incidence of the polymorphism rs2228014 of the CXCR4 gene (P = 1 and P < 0.05), as well as between the age of the patients and the simultaneous occurrence of these 2 polymorphisms (P = 0.20 and P < 0.05; Table 3).
Table 3. Relationship between some clinical data of patients with incidence and genotype incidence percentages
| P value | CXCL12 genotypes (rs1801157) (n = 30, %) |
CXCR4 genotypes (rs2228014) (n = 30, %) |
Clinical information of patients | ||||
| AA | AG | GG | TT | CT | CC | ||
| 0.0707 |
1(3/33) | 9(29/97) | 9(29/97) | 0 | 0 | 19(63.33) | Risk before menopause |
| 0 | 9(29/97) | 2(6/66) | 0 | 0 | 11(36.66) | Risk after menopause | |
| 0.899 |
0 | 1(3.33) | 3(9/99) | 0 | 0 | 4(13/32) | BMI (20-22 kg/m2) |
| 0 | 7(23/31) | 3(9/99) | 0 | 0 | 10 (33/3) | BMI (22-25 kg/m2) | |
| 1(3/33) | 4(13/32) | 1(3/33) | 0 | 0 | 6(19/98) | BMI (25-27 kg/m2) | |
| 0 | 5(6/65) | 2(6/66) | 0 | 0 | 7(23/31) | BMI (27-30 kg/m2) | |
| 0 | 2(6/66) | 1(3/33) | 0 | 0 | 3(9/99) | BMI (30-40 kg/m2) | |
| 0.020 |
0 | 6(20) | 7(23.33) | 0 | 0 | 13(43.33) | Age 50> |
| 1(3.33) | 13(43.33) | 3(10) | 0 | 0 | 17(56.66) | Age 50 = < | |
| 0.001062 |
0 | 1(3/33) | 0 | 0 | 0 | 1(3/33) | Metastasis 1> year |
| 0 | 3(9/99) | 1(3/33) | 0 | 0 | 4(13/33) | Metastasis 1-2 years | |
| 0 | 7(23/31) | 4(13/32) | 0 | 0 | 11(36/66) | Metastasis 3-4 years | |
| 1(3/33) | 5(16/65) | 5(16/65) | 0 | 0 | 11(36/66) | Metastases 5-6 years | |
| 0 | 2(6/66) | 1(3/33) | 0 | 0 | 3(9/99) | Metastasis 6< years | |
| 0.684 |
0 | 4(13/33) | 1(3/33) | 0 | 0 | 5(16/66) | Grade II tumor |
| 1(3.33) | 11(36/66) | 7(23/33) | 0 | 0 | 19(63/33) | Grade III tumor | |
| 0 | 4(13/33) | 2(6/66) | 0 | 0 | 6(20) | Grade IV tumor | |
Discussion
This study evaluated the data obtained over the years on the role of the CXCL12/CXCR4 signal in breast cancer and its potential use to develop new therapeutic tools for managing breast cancer and prognostic improvement. Therefore, in the present study, the relationship between rs2228014 and rs1801157 SNPs and metastatic breast cancer, the role of these genetic changes with clinical factors, and disease prognosis were investigated and evaluated in the patient and control groups.
Several studies have investigated the association between the rs1801157 polymorphism of the CXCL12 gene and the risk of various types of cancer, including breast cancer (17, 18). For instance, de Oliveira et al (2011) concluded that patients with breast cancer that were estrogen positive and carrying the A allele in the rs1801157 polymorphism showed less expression of CXCL12 (about 1.2 times) than the GG genotype in the blood cell (P = 0.53) (19).
Further, Schimanski et al (2011) showed that the presence of the A allele (AA and GA genotypes) in the rs1801157 polymorphism had a significant correlation with gastric and esophageal cancer metastases (P = 0.026), but it was not a diagnostic factor (20). Chen et al (2015) examined the relationship between CXCR4 rs2228014 and CXCL12 rs1801157 polymorphisms and the outcome of treatment in 222 patients with nasopharyngeal carcinoma (NPC). They found no significant relationship between the presence of both polymorphism and clinical factors. However, their analysis showed that the clinical stage and CXCL12 rs1801157 polymorphism were significantly associated with distant metastasis, disease-free survival, and progression-free survival (P = 0.018, 0.028, and 0.013). Their results showed that the CXCL12 rs1801157 AA genotype might act as a potential prognostic factor in NPC patients (21).
Moreover, Chang et al (2009) conducted a study on 102 patients with hepatocellular carcinoma (HCC). They showed that between SDF-1 (rs1801157) and CXCR4 (rs2228014) polymorphisms, only 1 significant relationship was between the susceptibility to the SDF-1 polymorphism (rs1801157) and HCC (22). Consistent with their results, in our study, the allele frequency of polymorphisms in both groups showed no significant relationship between the incidence of the A allele and breast cancer (P = 0.91). According to studies on the relationship between the presence of both polymorphisms and the incidence of breast cancer, no significant correlation was found between the simultaneous occurrence of normal and mutant alleles in both groups for rs1801157 and rs2228014 polymorphisms (P = 0.95 and P < 0.05).
There was also a significant relationship between the time of recurrence (metastasis) and the occurrence of CXCL12 rs1801157 and CXCR4 rs2228014 polymorphisms (P = 0.001 and P < 0.05). Also, many studies on the CXCR4 rs2228014 polymorphism have shown that this polymorphism is only effective in lung cancer and chronic myeloid leukemia (23-25). Kishima et al (2015) studied the effects of CXCR4 polymorphisms on breast cancer, showing that the genetic variants of CXCR4 rs2228014 have no effect on the protein or expression of CXCR4 in breast cancer (P = 0.30), although this protein is much more expressed in breast cancer patients than normal individuals (P = 0.75) (26). Lee et al (2011) showed a significant correlation between CXCR4 rs2228014 and CXCL12 rs1801157 polymorphisms and the potential for lung cancer and the detection of non–small cell lung cancer. In this study, people with the AA genotype in CXCL12 (P = 0.018 and P < 0.0001) or TT genotype in CXCR4 (P = 0.002 and P < 0.0001) were more susceptible to acute or poorer diagnosis than other people (23). In our study, regarding the peripheral blood of patients, the incidence of the rs2228014 polymorphism in the CXCR4 gene was not significantly different between the 2 groups (P = 1).
In a recently published review article, Yazdani et al focused on the role of the CXCL12/CXCR4 axis in various aspects of acute myeloid leukemia (AML) cancer. They indicated that compounds that interfere with the CXCL12/CXCR4 axis targeted both leukemic and stromal cell interaction as an attractive strategy for improving the outcome of treatment in AML (24). In our study, some clinical features of the patients were also examined. According to the results, there was a significant relationship between age and the incidence of CXCL12 rs1801157 polymorphism (P = 0.02), and there was a close relationship between menopausal age and disease incidence (P = 0.07). According to most studies, the incidence of breast cancer is associated with being overweight. The risk level is 1.5-2 times more in obese women than in others, and this risk is related to the period of menopause (25, 27). Also, a higher incidence of menopause is associated with an increased risk of breast cancer (28). On the other hand, we found that factors such as age, BMI, and tumor grade were not significantly correlated with the rs2228014 (CXCR4) polymorphism; also, we did not find a relationship between the rs1801157 (CXCL12) polymorphism and tumor grade. It was observed that there was no significant relationship between tumor grade and the occurrence of rs1801157 (CXCL12) and rs2228014 (CXCR4) polymorphisms. According to the findings of this study, the studied polymorphisms cannot be considered a definite prognosis for breast cancer. It should be noted that the limited number of studied samples affected the results of this research. However, the difference in the abovementioned results that could be due to various genetic patterns in different parts of the world (as well as the difference in the number of the examined samples) was not unexpected. However, to achieve definitive findings, further studies are needed to be conducted on other ethnic groups of Iran with larger populations.
Conclusion
No significant correlation was found between the incidence of mutant alleles in CXCR4 (rs2228014) and CXCL12 (rs1801157) polymorphisms and the incidence of the disease. However, there was a significant relationship between chemotherapy and the occurrence of these polymorphisms in women with metastatic breast cancer under chemotherapy in Zanjan. Also, some of the studied factors (including the age of menopause and the age of the patients) were associated with the disease. Different results were obtained due to differences in genetic and racial patterns in the studied populations, the number of studied subjects, genetic drift, etc. Further studies are needed to study the relationship between these polymorphisms, metastatic breast cancer, and the therapeutic outcome of patients.
Acknowledgements
This article is part of a master’s thesis (research number: 62630503942023) approved by the Graduate Council of Islamic Azad University of Zanjan. The authors are grateful to the staff of the Biology Research Center of Zanjan Azad University and the staff of the Chemo Department of Vali-e-Asr Hospital because of their close cooperation in this project.
Conflicts of Interest
The authors have no conflicts of interest to be declared.
Received: 2022/01/22 | Accepted: 2022/09/20 | Published: 2023/03/13
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |