BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://journal.zums.ac.ir/article-1-6307-en.html


 , Rasoul Yousefimashouf1
, Rasoul Yousefimashouf1 

 , Iraj Sedighi2
, Iraj Sedighi2 

 , Abbas Moradi3
, Abbas Moradi3 

 , Fatemeh Nouri4
, Fatemeh Nouri4 

 , Mohammad Taheri *5
, Mohammad Taheri *5 


2- Dept. of Pediatric, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran.
3- Dept. of Community Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
4- Dept. of Pharmaceutical Biotechnology, School of Pharmacy, Hamadan University of Medical Sciences, Hamadan, Iran
5- Dept. of Medical Microbiology, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran ,
✅ The afa and sfa genes have a significant correlation with strong biofilm formation in uropathogenic E. coli (UPEC).
Urinary tract infection (UTI) in children is one of the most frequent infections, and it has a significant economic cost (1). Uropathogenic Escherichia coli (UPEC) is one of the most prevalent pathogens that cause UTI and asymptomatic bacteriuria in children (2, 3). It causes a variety of problems in children of all ages, but because of anatomical differences in the urinary system, it is more common in girls (4). The severity of a UTI is determined by bacterial virulence factors and human immunity. Fimbriae, toxins, and iron absorption systems are the most important factors of bacterial virulence that can play a role in biofilm formation (5). UPEC to bind to epithelial cell receptors can produce several adhesion types of H and S fimbriae (6, 7). S fimbriae, which interacts with sialic acid in the bladder epithelium and urethral ducts, is encoded by the sfa gene (8, 9). Biofilm production is one of the pathogenic processes that allows E. coli to remain in the urinary system and colonize it more effectively (10). Bacterial biofilm is a cluster of bacteria encased in a polysaccharide matrix that lowers antibiotic sensitivity. They also participate in the interchange of food and genetic material such as plasmids from one bacterium to another, and the emergence of antibiotic resistance (11). Because of enhanced resistance to antimicrobials and the host immune system, the formation of microbial biofilms poses a significant obstacle to the progression of the illness (12). Antibiotic resistance is on the rise among members of the Enterobacteriaceae family, notably E. coli, in children all around the world. Antimicrobial resistance in E. coli decreases treatment options, increasing healthcare expenses and mortality (13). Antibiotic administration patterns in various geographical locations can have a role in the development of antibiotic resistance in that area (14).
Due to the high prevalence of UTI in children and the significance of biofilms in the development of this illness, the aim of current study was to explore the link between biofilm formation in isolated UPEC strains and its antibiotic resistance pattern in patients with UTI in Hamadan, Iran.
Sample preparation
In the current cross-sectional study, 112 E. coli strains isolated from pediatrics with UTI (105 CFU/mL of urine) referred to Besaat and Sina hospitals. Bacterial strains were isolated from November 2018 to April 2019. The subjects in this study were outpatients who had UTIs; bacterial growth of E. coli isolated from urine culture was judged positive. Exclusion criteria were the use of a urinary catheter, consumption of antibiotics during the previous two weeks, and a urinary tract disorder. Morphological and biochemical testing was used to confirm the specimens. They were incubated at 37 °C overnight and confirmed with biochemical differentiation tests. The bacterial isolates were stored at -20 °C (15).
Antimicrobial Susceptibility Testing
Antibiotic susceptibility was carried out via the Kirby-Bauer disk diffusion assay. All antibiotic discs were purchased from ROSCO, Denmark. The standard 0.5 McFarland microbial suspension was prepared according to CLSI 2018 for antimicrobial susceptibility testing.
Antimicrobial susceptibility tests were done on Muller-Hinton agar after the 0.5 McFarland bacterial suspension was prepared. As a standard control, E. coli ATCC 25922 was applied. The antibiotic discs used in the study were Ciprofloxacin (5 μg), Nitrofurantoin (300 μg), Meropenem (10 μg), Imipenem (10 μg), Cephalothin (30 μg), Cefotaxime (30 μg), Nalidixic acid (30 μg), Cefixime (5 μg), Ceftriaxone (30 μg) Cotrimoxazole (trimethoprim 1.25 μg + sulfamethoxazole 23.75 μg), Amikacin (30 μg) and Gentamicin (10 μg) (15, 16).
Biofilm Formation
Biofilm production ability in UPEC isolates was examined by the microtiter plate. The assay was performed in triplicate. E. coli isolates were prepared and incubated at 37 °C overnight. A 0.5 McFarland standard microbial suspension was prepared; 200 µL of was added to each well and incubated for additional 16-18 hours at 37 °C. Normal saline was used three times in the washing step, then to fix cells, 200 µL absolute ethanol (96%) was added to wells; the good contents were pulled after 15 minutes, and lets plate was dried at room temperature. The staining step was carried out by adding 200 µL of 2% crystal violet for five minutes. After the color was removed, 200 µL of 33 percent acetic acid was added to each well and incubated for 15 minutes at 37 °C; the optical density was then recorded using an ELISA Plate Reader at a wavelength of 492 nm (17, 18).
DNA Extraction
Total DNA extraction was performed using a boiling method. E. coli bacteria were first dissolved in 300 µL of distilled water and boiled for 10 minutes at 95 °C. Centrifugation was performed at 12,000 rpm for 10 minutes. 100 µL of supernatant is considered a DNA template. DNA concentration was checked through a nanodrop spectrophotometer. In the present study, PCR detected afa and sfa genes. The specific primer sequences have been shown in Table 1.
Table 1: Primer sequences and annealing temperature used for afa and sfa genes amplification
| Genes | Primer Sequence | Fragment size (bp) | annealing temperature | Reference |
| afa | F:5ʹ-CGGCTTTTCTGCTGAACTGGCAGGC-3ʹ R:5ʹCCGTCAGCCCCCACGGCAGACC3ʹ |
672 bp | 59 | 15 |
| sfa | F;5ʹ-CTCCGGAGAACTGGGTGCATCTTAC-3ʹ R:5ʹ-CGGAGGAGTAATTACAAACCTGGCA-3 |
410 bp | 63 | 15 |
Polymerase Chain Reaction Test
PCR reaction mixture was prepared in a total volume of 20 µL including 10 µL of master mix, 1 µL of forward and reverse primer (10 pmol/L), 1 µL of DNA template, and 3.5 µL of nuclease-free water. In the present study, standard strains harboring afa and sfa genes were utilized as the positive control to validate the accuracy of each test. Gene amplification was done in a thermocycler (Bio-Rad, USA); the PCR program including initial denaturation, one cycle at 95 °C for one minute, followed by 35 cycles; denaturation at 95 °C for 30 seconds; annealing at 59 °C and 63 °C for afa , and sfa genes respectively, for 45 seconds; extension, 72 °C one minute, and the final extension at 72 °C for 5 minutes (22). The PCR products were separated by electrophoresis on a 1% agarose gel (Merck Germany) and visualized using Gel Documentation (Uvitec, UK).
Statistical Analysis
SPSS Statistics version 17.0 was used to analyze the data (SPSS Inc., Chicago, Ill., USA). The chi-square and Fisher's exact tests were used to compare nominal variables represented as mean or frequency. Two-tailed probability was used and the p-values less than 0.05 were considered statistically significant.
The present study was performed on 112 E. coli strains in Hamadan from 2018 to 2019. The Age range of the children with UTI was one month to 12 years (mean 5.881). Regarding gender, 29 (25.9%) and 83 (74.10%) isolates were isolated from boys and girls, respectively.
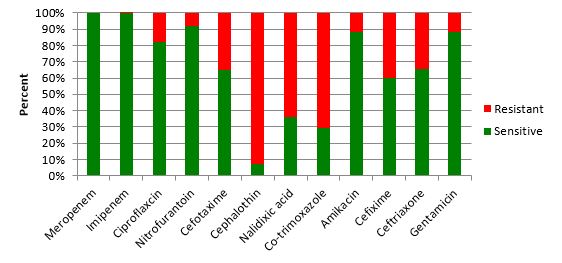
Antibiotic susceptibility and resistance pattern (Figure 1) revealed that the highest resistance was found against Cephalothin (76.79%), and the lowest resistance was observed to meropenem and imipenem (0%). There was no significant association between gender and biofilm production (p ≥ 0.05). The results of microtiter plate method in our research indicated that 81 (72.32%) E. coli strains isolated from UTI patients were biofilm producers. Based on OD, biofilm-production capacity was classified as high, moderate, or weak. Of the 81 biofilm-producing E. coli strains, 18 (22.22%) were strong, 27 (33.33%) moderate, and 36 (44.44%) weak biofilm producers. Nevertheless, 31 (27.67 %) isolates did not show biofilm formation. The production of strong biofilms and sfa genes were found to have a substantial relationship (p < 0.0001).
Figure 1: Antibiotic susceptibility testing of uropathogenic Escherichia coli isolates.
In the current study, the relationship between biofilm production and antibiotic resistance in antibiotics such as Nitrofurantoin and Cefotaxime was statistically significant (p < 0.05). Nevertheless, the relationship between biofilm production and other antibiotics was not statistically significant (p ≥ 0.05).
Regarding the frequency of virulence genes in 112 E. coli isolates by PCR, 33 strains (29.4%) harbored afa gene, and 55 (49.1%) isolates carried the sfa gene. The results of PCR amplification of afa and sfa gene in UPEC isolates are represented in Figures 2 and 3, respectively. There was a statistically significant (p < 0.039) relationship between MDR and afa gene frequency. Even though there is no significant relationship between MDR and biofilm production (p ≥ 0.05).

Figure 2: PCR amplification of afa gene in UPEC isolates. DNA Ladder (50 - 1000 bp); Lane 1, positive control; Lanes 2-4 and 7, PCR product from afa gene; Lane 5, negative control.
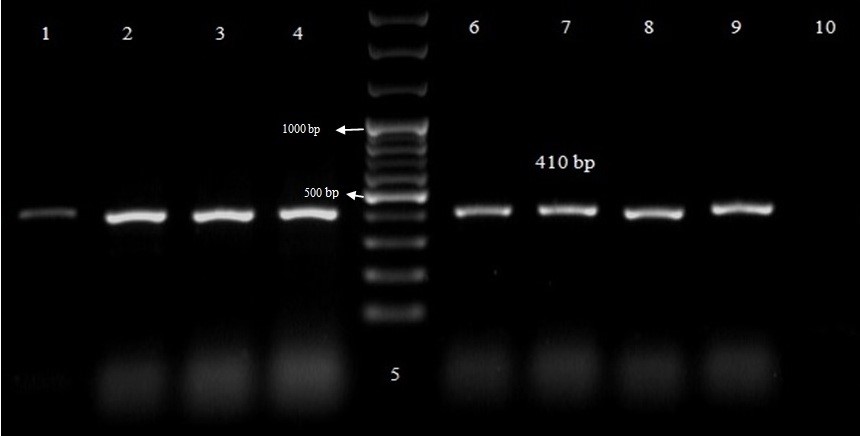
Figure 3: PCR amplification of sfa gene in UPEC isolates. Lanes 1-4 and 7-9, PCR product from sfa gene; DNA Ladder (50 - 1000 bp); Lane 6, positive control; Lane 10, negative control.
Discussion
Among the bacteria that cause urinary tract infections, E. coli is one of the most significant and common agents in children. In the current study, the frequency of UPEC in girls was higher than in boys, which could be attributed to female urinary tract anatomical conditions (19). This result contrasts with the findings of a study by Sedighi et al., i.e., the sensitivity of Imipenem (96.7%). Susceptibility to Imipenem was expected to decline over time in this study. In recent years, increased sensitivity can be attributed to various protocols in studies and other antibiotic disks (20, 21).
According to overuse of antibiotics, such as Co-trimoxazole, it is shown that UPEC bacteria were resistant to Cotrimoxazole is increasing from 2000 to 2010, so that 6.3% increasing rate was reported (10). This increase in antibiotic resistance in this study could be attributed to excessive consumption of antibiotics, food intake from antibiotic-containing animals, the transmission of resistance isolates between individuals, self-medication, and non-compliance with conventional antibiotics (16,17). It is believed that microbial adherence is the main critical stage involved in the pathogenicity of E. coli to the uroepithelial cells. The fimbriae handle this connection and colonization (22, 23). The investigations of P, S, f1s fimbriae and understanding microbial pathogenesis could assist physicians in predicting infection management. To determine the prevalence of virulence factors in E. coli, several molecular studies were conducted (20, 24).
Biofilm formation plays a key role in the increment of nosocomial infections and UTI in children. The literature review showed that biofilm-forming bacteria are responsible for more than 50% of bacterial infections (2, 25). In the present study, of the 81 biofilm-producing E. coli strains, 18 (22.2%) were strong, 27 (33.3%) moderate, and 36 (44.4%) weak biofilm producers. According to a study by Kiehlbauch et al., 18%, 25%, and 56% of E. coli strains produced strong, moderate, and weak biofilms, respectively (24). Based on the study by Poursina et al., 80% of all UPEC isolates were biofilm producers, of which 29%, 34%, and 17% had strong, moderate, and weak biofilm-producing ability, respectively (26). However, in our study, the frequency of biofilm formation is 22.2% strong, 33.3% moderate, and 44.4% weak; that this difference can be due to different geographical regions. Boroumand et al. reported that 75.38% of E. coli strains were biofilm producers (27). This high frequency can be due to genetic factors in the bacteria, and the use of certain groups of antibiotics. In Zamani study, among E. coli strains 36%, 48%, and 10% showed high, moderate, and weak biofilm production, respectively and the rest of them were non-biofilm producers (28). Mittal et al. reported that 13.5% of E. coli were biofilm producers (29). Ponnusamy et al. recognized all isolated strains as biofilm-forming bacteria, with 6.6%, 80.8%, and 14.14% creating strong, moderate, and weak biofilms, respectively (30). Besides, Neupane et al. reported an increase in biofilm production among 51.92% E. coli strains using Congo Red Agar CRA (35). In a study by Ren et al., the biofilm development of 116 E. coli strains was examined. Biofilm was formed by 41.3% of the strains. 16.4%, 18.1%, and 6.8% formed weak, moderate, and strong biofilms, respectively (31). Variation in biofilm formation outcomes could be attributable to geographic disparities in low sanitation, varied antibiotic resistance, and different origin of isolates in patients with urinary tract infections. According to our results, the frequency of afa and sfa adhesive genes in E. coli were 24.9% (n = 33) and 49.1% (n = 55), respectively. In the current study, high frequency related to S fimbriae of E. coli indicated the importance of this virulence factor in the development of UTI. Furthermore, the prevalence of fimbriae S suggested that this factor was a fimbria requirement for adhesion and colonization E. coli in UTI (22, 32). Several similar studies have been done in different parts of the world, the results of which are different and can be due to differences in geographical areas, how antibiotics are prescribed, and the genetics of the disease-causing microorganisms (28, 33, 34). The current study results showed that strains isolated from biofilm structure revealed a significant incidence of afa and sfa genes. The afa and sfa genes were found in 92.85% and 85.71 percent of high biofilm-producing isolates. The results of our study indicated that sfa gene in UPEC has a substantial connection with high biofilm development (P-value < 0.0001). In this study, the frequency of afa and sfa adhesion genes were 29.4% and 49.4% respectively, which showed the important role of adhesins in UTI pathogenesis. S fimbria as a virulence factor in UPEC isolates can bind to the epithelial cells of the urinary tract system in humans and is a pivotal parameter in the biofilm production of UPEC strains (31, 35).
Investigation of the frequency of genes involved in bacterial adhesion can be effective in identifying and changing the medical approach. The monitoring of genes that are directly related to biofilm formation and antibiotic resistance can help researchers and physicians in the prevention, control, and treatment of infectious diseases.
Conclusion
The afa gene was shown to be more prevalent than sfa in E. coli strains. Strong biofilm development was linked to both the afa and sfa genes. Furthermore, the biofilm formation rate was higher in antibiotic-resistant E. coli strains than in antibiotic-susceptible bacteria. To avoid the establishment of multi-drug resistant bacteria, it is recommended that UTI in Hamadan be treated with Imipenem or Meropenem, based on the antibiotic resistance pattern. Other virulence factors involved in biofilm formation in E. coli that cause UTI should be investigated. Amikacin, Gentamicin, and Nitrofurantoin antibiotics can additionally be suggested to treat UTI as an empirical prescription in this geographical area. In this study, it was also found that sfa gene has a critical role in the pathogenesis of E. coli isolated from urinary tract infections in children.
Acknowledgements
The authors thank all those who helped them writing this article.
Conflicts of Interest
There is no conflict of interest.
Received: 2020/11/26 | Accepted: 2021/03/3 | Published: 2021/10/17
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |