BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://journal.zums.ac.ir/article-1-6402-en.html


 , Ryhaneh Babaei2
, Ryhaneh Babaei2 

 , Maryam Jalali2
, Maryam Jalali2 

 , Hamid Reza Goli2
, Hamid Reza Goli2 

 , Mohammad Reza Haghshenas2
, Mohammad Reza Haghshenas2 

 , Negin Hosseini3
, Negin Hosseini3 

 , Ebrahim Shafaie4
, Ebrahim Shafaie4 


2- Dept. of Medical Microbiology and Virology, Faculty of Medicine, Mazandaran University of Medical Sciences, Sari, Iran
3- Dept. of Medical Microbiology and Virology, School of Medicine, Zanjan University of Medical Sciences, Zanjan, Iran
4- Infectious Diseases Research Center, Birjand University of Medical Sciences, Birjand, Iran
✅ Given the bacterial isolates detected from hospital wards and following the findings prompt diagnosis method is essential to control infections and the proper use of effective antibiotics.
Nosocomial infections are the infections that affect hospitalized patients during their stay in the ward and patients symptoms might appear either when they are being hospitalized and/or after the patients are discharged (1).
Bacteria are found abundantly in the media particularly in hospitals (2). These are often opportunist pathogens and are able to generate various types of hospital-acquired infections such as bacteremia, meningitis, respiratory system infection, urinary tract infection, surgery wound infections and so on (3, 4). The importance of bacteria in medical centers could be discussed from many aspects. Due to the antibiotic resistance, the members of this genus increases in an accelerated rate so far that currently, with the development of highly resilient strains, treating bacteria-generated infections has been facing challenges (5). Meningitis has been noted as an overwhelming disorder with high mortality and morbidity rates (170000 case per year) (6). Blood infection is the reaction of the defense system in the body against infectious agents such as bacteria, viruses, or fungi. In some cases, Pseudomonas aeruginosa (P. aeruginosa) is isolated from blood infections as well (7). Nosocomial infections might cause meningitis, i.e., inflammation of meninges in peripheral areas of the brain and spinal cord. Despite antibiotic treatment, bacterial meningitis causes extensive damages and mortality. Studies conducted in Iran showed that Streptococcus pneumoniae (S. pneumoniae) was respon-sible for 30% of meningitis cases, Haemophilus influenza (H. influenza) for 15%, Coagulase-negative staphyloco-cci for 14% and Neisseria meningitides (N. meningitidis) for 13%. Generally, nosocomial infections not only increase the costs due to prolonged hospital stay,and going through diagnostic and treatment measures for detecting the routes of microorganisms transmission in hospital environment; they also cause high mortality and illness rate.
Regarding the essential impact of bacteria in bacteremia and meningitis; and the necessity to improve knowledge on the prevalence of developing the diseases and identifying bacterial agents related to such cases, in order to facilitate infection control in certain hospitals, the present study aims at assessing the presence of bacteria and their susceptibility patterns against antibiotics.
In this cross-sectional study, the bacterial isolates of the two laboratories in Imam Khomeini and Bouali Hospitals of Sari, Iran, were studied from April 2016 to March 2018.
Samples including morning midstream urine, blood, stool, wound discharge, cerebro-spinal fluids (CSF), and respiratory specimens were aseptically collected by sterile containers and carried to the laboratory as soon as possible . Bacterial isolates were identified via morphologic features including colony specification, gram staining, and standard biochemical diversity following standard methods (8).
Clinical specimens collection:
Detecting the bacterial specimens:
Detecting anaerobic bacteria:
Susceptibility testing:
The discs which were used in Gram positive bacteria were gentamycin (10 μg), erythromycin (15 μg), ciprofl-oxacin (5 μg), clindamycin (15 μg), oxacillin (1 μg), and, vancomycin (30 μg) for genus staphylococci. Methicillin (5unit), erythromycin (15 μg), gentamycin (10 μg) and clindamycin (15 μg) were used for pneumococci. In addition, for Gram negative bacteria, ampicillin (10 μg), co- amoxiclav (500 μg/125 μg), ceftazidime (30 μg), imipenem (10 μg) gentamycin (10 μg), ciprofloxacin (5 μg), cephalexin (30 μg), co-trimoxazole (23.75+1.25 μg) and cefazolin (30 μg) for P. aeruginosa; and, ampicillin (10 μg), cefepime (30 μg), ceftriaxone (30 μg), ceftazi-dime (30 μg), cefixime (5 μg), imipenem (10 μg), nalidi-xic acid (30 μg), gentamycin (10 μg) and ciprofloxacin (5 μg) were used for Enterobacteriaceae genus. (All antibiotics were supplied from Patanteb Iran). The antibiotic resistance pattern of the bacteria isolated from clinical specimens was defined by using the disk diffusion test in Kirby–Bauer method based on Clinical Standards Institute and CLSI (13). E. coli ATCC 25922, K. pneumoniae ATCC 7881, S. epidermidis 12228 and S. aureus ATCC 43300 were used as positive controls.
Statistical Analysis:
In this study, we used the SPSS software version 20 for the analysis of collected data. The Exact Fisher test was used for assessing the relationship between qualitative variables if needed and the p value<0.05 was considered as the significant level.
Ethical Statement:
The ethical statement IR.MAZUMS.REC.1397.1296 was the criterion for performing all stages of this study.
Of the 634 positive specimens recovered from Bouali and Imam Hospitals, Escherichia coli (E. coli) (39.4%), Staphylococcus epidermidis (S. epidermidis) (17.8%) and other Coagulase –negative staphylococci (11%) were the most common pathogens found in the recovered speci-mens. Those three microorganisms constitute around two thirds (68.2%) of total pathogens. (Fig.1).
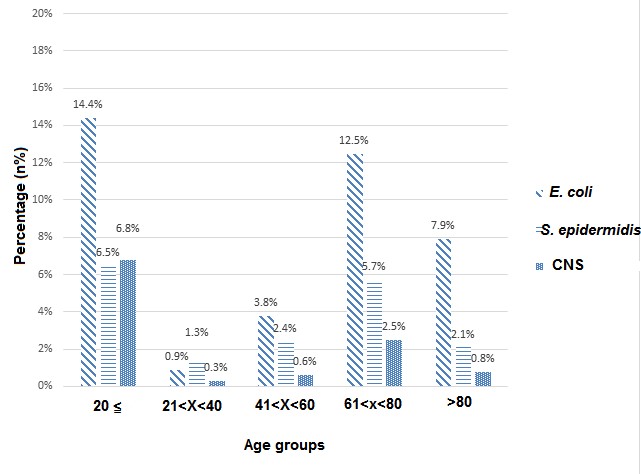
Figure 1. The percents of frequency of microorganisms: E. coli, S. epidermidis and Coagulase- negative Staphylococci were among the most common microorganisms in the whole specimens (as per age group at Boali and Imam Khomeini Hospitals).
In general, among the 634 cases, 420 (66.2%) urine, 202 (31.9%) blood and 12 (1.9%) CSF specimens were evaluated.
The most common frequency of the specimen recovered from intensive care unit (ICU) was 16.6%, followed by internal wards (15.9%), neurology (14.5%), infection, allergy and pediatrics (10.4%). The ophthalmo-logy ward (0.5%), ENT. (1.1%) and men and women’s ward (1.6%) had the least prevalence in terms of frequency of culture specimen. In general, in the internal diseases, neurology, ICU, ER, infectious, children allergy, Surgery, Neonatal Intensive Care Unit (NICU), Pediatrics and ENT, E. coli was the most prevalent pathogen agent in hospital-acquired infections. In the neonates ward, S. epidermidis and coagulase- negative staphylococcus coagulase were the most common pathogens. In PICU Ward, pseudomonas aeruginosa was the most common pathogen. In oncology, ophthalmology, male and gyneco-logy wards, S. epidermidis was the most common microorganism responsible for nosocomial infections. Enterococci was detected only in one (1) case and it was only in emergency ward. Streptococcus viridance was detected only in three (3) patients, two (2) were inpediatric ward and one (1) case was in the internal ward. Serratia was identified only in two patients in surgery ward. H. influenza was observed in two cases (one in oncology and one in surgery ward), Candida was detected in the specimens extracted from 3 patients in ICU (Based on the hospitals laboratory reports), pediatric intensive cares (PICU) and Neonatal intensive care unit (NICU). Salmonella was detected in 2 wards, emergency and PICU. Proteus was found in two ENT, male and females wards. In general, no statistically significant difference was found between microorganisms and the hospital wards (P= 0.298).
From urine cultures E. coli (37.5%), S. epidermidis (14.2%) and P. aeruginosa (4.7%) were isolated. The main responsible bacteria for bacteremia were coagulase –negative staphylococci (9.8%), S. epidermidis (3.5%), and P. aeruginosa (3.5%); respectively. Coagulase- negative staphylococci, S. pneumoniae and H. Influenza were the most meningitis-related isolates.
In the meningitis, the Coagulase- negative Staphylo-cocci specimens were completely sensitive to, ciproflo-xacin and Gentamycin while they were resistant to Methicillin 100%. S. pneumoniae was sensitive to amikacin and Gentamycin. No drug resistance was seen in H. Influenza. S. epidermidis was completely sensitive to co-amoxiclav, co-trimoxazole, gentamycin, oxacillin and vancomycin; on the other hand, it showed resistance to ciprofloxacin. In Bacteremia E. coli, the specimens were completely sensitive to amikacin, cefotaxime, ciprofloxacin and Nitrofurantoin; while, they were resistant against amoxiclav and nalidixic acid. In general, E. coli, S. epidermidis, P. aeruginosa and Coagulase- negative Staphylococci were completely sensitive to amikacin; and, Klebsiella spp and S. aureus showed 93.3% sensitivity to amikacin. Ampicillin had 100% sensitivity against S. epidermidis; however, P. aerugi-nosa, Klebsiella and Acinetobacter showed more than 90% resistance against ampicillin. E. coli, Klebsiella spp and S. aureus showed 100% resistance to amoxiclav; while Klebsiella sp was completely sensitive to amoxiclav. Acinetobacter was resistant against ceftria-xone. S. epidermidis, P. aeruginosa, Klebsiella spp, Coa-gulase- negative Staphylococci, S. aureus and Acine-tobacter were highly resistant against cephalexin. S. epidermidis, K. pneumoniae and Acinetobacter ishowed full resistance to Ceftazidime. P. aeruginosa was resistant against cefazolin, clindamycin and nitrofurantoin. Of the total 202 cases of positive blood culture specimens, there were only two pathogens responsible for bacteremia symptoms in the Neurology Ward which consisted of Coagulase-negative Staphylococci and Klebsiella sp. (Table 1- 2). (Susceptibility testing of G+ and G-isolated was listed in Table 3, 4).
Table 1. The frequencies of specimens associated with bacteremia, meningitis and urinary tract infections in two target hospitals regarding the wards from which the specimens were taken.
| Hospital ward | Bouali Hospital | Imam Khomeini Hospital | ||||
| Urinary tract infection | Bacteremia | Meningitis | Urinary tract infection | Bacteremia | Meningitis | |
| Internal & Gynecology | 87 (20.7%) | 12 (5.9%) | 2 (16.7%) | 101 (25.3%) | 43 (28.6%) | - |
| Neurology Surgery | 88 (21%) | 2 (1%) | 2 (16.7%) | 85 (21.3%) | 15 (10%) | 12 (60%) |
| ICU | 64 (15.2%) | 41 (20.3%) | 2 (16.7%) | 88 (22%) | 28 (18.7%) | 8 (40%) |
| Emergency | 56 (13.6%) | 9 (4.5%) | - | 61 (15.3%) | 64 (42.7%) | - |
| Total Specimens | 420 (100%) | 202 (100%) | 12 (100%) | 400 (100%) | 150 (100%) | 20 (100%) |
Table 2. Frequencies of antibiotic-resistant bacterial isolates in the two target hospitals.
| Antibiotic | Boali Hospital | Imam Khomeini Hospital | ||||
|---|---|---|---|---|---|---|
| Urinary tract infection | Bacteremia | Meningitis | Urinary tract infection | Bacteremia | Meningitis | |
| Amikacin | 18 (4.3%) | 8 (4%) | 1 (50%) | 42 (10.5%) | 85 (56.7%) | 14 (70%) |
| Ampicillin | 252 (60%) | 74 (36.6%) | 3 (100%) | 240 (60%) | 63 (42%) | 7 (35%) |
| Ceftriaxone | 128 (30.5%) | 76 (37.6%) | - | 253 (63.2%) | 57 (38%) | 1 (5%) |
| Cefixime | - | - | - | 262 (65.5%) | - | - |
| Ceftazidime | 79 (18.8%) | 26 (12.9%) | - | 124 (31%) | 61 (40.7%) | - |
| Cefotaxime | - | 41 (20.3%) | 2 (22.2%) | 138 (34.5%) | - | 4 (20%) |
| Cefazolin | - | 24 (11.9%) | 2 (40%) | 245 (61.3%) | 97 (64.7%) | - |
| Ciprofloxacin | - | 22 (10.9%) | 1 (14.3%) | 241 (60.3%) | 69 (46%) | 14 (70%) |
| Co-trimoxazole | 51 (12.1%) | 44 (21.8%) | 2 (28.6%) | 278 (69.5%) | 103 (68.7%) | - |
| Gentamycin | 72 (17.1%) | 44 (21.8%) | 2 (25%) | 66 (16.5%) | 33 (22%) | 12 (60%) |
| Imipenem | 61 (14.5%) | 24 (11.9%) | - | 72 (18%) | 56 (37.3%) | 16 (80%) |
| Nalidixic acid | 152 (36.2%) | - | - | 322 (80.5%) | - | - |
| Nitrofurantoin | - | - | - | 117 (29.3%) | - | - |
| Vancomycin | 9 (2.1%) | 8 (4%) | - | 35 (8.8%) | 3 (2%) | 0 (0%) |
Table 3. Antibiotic resistance in Gram Positive isolates. Some antibiotic susceptibility was done based on the empirical therapy.
| Staphylococcus epidermidis | Staphylococcus saprophyticus |
Staphylococcus aureus | Other Coagulase negative Staphylococci | Streptococcus pneumoniae | Viridans Streptococci | Bacillus | |
|---|---|---|---|---|---|---|---|
| Ampicillin | resistant | - | - | - | - | - | - |
| Co-amoxiclav | sensitive | - | resistant | - | - | - | sensitive |
| Amikacin | sensitive | - | 93.3% sensitive |
sensitive | - | - | sensitive |
| Erythromycin | - | - | resistant | resistant | - | - | - |
| Oxacillin | sensitive | - | - | - | - | - | sensitive |
| Ceftazidime | resistant | sensitive | - | sensitive | sensitive | - | sensitive |
| Cefixime | - | - | - | sensitive | - | - | - |
| Cephalexin | resistant | - | resistant | resistant | - | - | - |
| Cefotaxime | - | - | - | - | - | sensitive | sensitive |
| Ciprofloxacin | resistant | - | - | sensitive | sensitive | - | - |
| Cefazolin | - | resistant 75% | - | - | - | sensitive | - |
| Gentamycin | sensitive | - | - | sensitive | sensitive | sensitive | - |
| Clindamycin | resistant | - | - | - | - | - | - |
| Co-trimoxazole | sensitive | - | - | - | - | resistant | sensitive |
| Methicillin | - | - | - | resistant | resistant | - | - |
| Nitrofurantoin | sensitive | - | - | - | - | - | resistant |
| Nalidixic acid | - | - | resistant | - | - | - | - |
| Vancomycin | 98.1% sensitive |
- | 80% sensitive |
92.3% sensitive |
- | sensitive | sensitive |
Table 4. Antibiotic resistance in Gram Negative isolates.
| Escherichia coli | Enterobacteriaceae | Acinetobacter | Proteus | Salmonella | Serratia | Pseudomonas Aeruginosa | Klebsiella | Klebsiella pneumoniae | Haemophilus Influenza |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | 82.3% resistant | - | resistant | - | resistant | resistant | resistant 97.7% |
resistant 97.4% | resistant 87.5% | resistant |
| Co-amoxiclav | resistant | _ | _ | - | - | - | - | resistant | sensitive | - |
| Amikacin | - | - | - | - | sensitive | sensitive | sensitive 97.7% | sensitive 78% | - | sensitive |
| Imipenem | - | - | - | sensitive | sensitive | sensitive | - | - | - | sensitive |
| Ceftazidime | - | - | resistant | - | - | sensitive | resistant 86.7% | - | resistant | - |
| Cefixime | resistant | - | - | - | - | - | resistant | - | resistant | - |
| Cephalexin | resistant | resistant | sensitive | resistant | resistant | resistant 95.5% | resistant | - | - | |
| Cefepime | resistant | - | - | - | - | - | resistant | - | - | - |
| Cefotaxime | sensitive | resistant | - | - | resistant | sensitive | - | - | - | - |
| Ceftriaxone | - | - | resistant | sensitive | - | sensitive | - | - | sensitive 73.3% | - |
| Ciprofloxacin | sensitive | resistant | - | - | sensitive | - | - | - | - | - |
| Cefazolin | - | - | resistant | - | - | - | resistant | - | - | - |
| Gentamycin | - | - | - | sensitive | - | sensitive | - | - | - | sensitive |
| Clindamycin | - | - | - | - | - | - | resistant | - | - | - |
| Co-trimoxazole | -- | - | sensitive | - | - | - | - | resistant 87.5% | - | - |
| Nitrofurantoin | sensitive | sensitive | - | - | sensitive | - | resistant | - | resistant | - |
| Nalidixic acid | resistant | - | - | - | - | - | - | - | - | - |
| Vancomycin | - | - | - | - | - | - | - | - | sensitive | - |
Discussion
Any organs of human body might show symptoms of infection in the hospital; however, among hospital-acquired infections, the urinary tract infection (42%), lower respiratory system infection or pneumonia (15% to 20%), infections associated with surgical wounds (24%) and infections of blood circulatory system (5%-10%) are of specific importance (14). In this study, out of 1024 clinical samples, frequencies of meningitis in two screened hospitals (Boali and Imam) were 1.9% and3.5% respectively, the amounts of bacterial rates to bacteremia were 31.9% and 26.3% as well. S. aureus (41.7%) and S. pneumoniae (45%) were the highest isolates in the mentioned hospitals for meningitis.
A published study in 2012 on 57112 hospitalized patients revealed that 592 of the patients had been affected by hospital-acquired infections. The rate of total prevalence of nosocomial infections was reported 1.03% which was mainly in the burns unit. The most common hospital-acquired infection was seen in the wound infections (44.6%); and the most common organisms were P. aeruginosa and Acinetobacter spp., (15). In our study, E. coli (39.4%), S. epidermidis (17.8%), Coagulase- negative Staphylococci (11%) are the rampant isolates found in the extracted specimen, which are similar to the Zahedi’s findings who reported the most common bacteria isolated in all specimen were E. coli (48.8%), S. epidermidis (22.9%) and K. pneumoniae (12%) (16), while in the studies conducted by Davoudi and her colleagues A. baumanni and P. aeruginosa were detected as the most common organisms (15). Based on the previously published findings, higher bacterial isolates related to the meningitis were N. meningitis (43%) ,H. influenza (13.6%). K. pneumoniae (13.7%) and S. aureus (11.1%) were detected as lower bacterial isolated rates in the mentioned study (6). At the current research out of 634 positive samples, S. aureus (41.7%) and S. pneumoniae (45%) were the highest isolates in the mentioned hospital for meningitis. The number of clinical speciemens, and geographical distributions of sample source are mentioned as the difference reasons.
Development of resistance to antibiotics in the pathogen bacteria poses one of the medical challenges across the world. This problem is more considerable in countries with irregular and unreasonable antibiotic consumption (17). In our study, Coagulase-negative Staphylococci and S. pneumoniae were the most meningitis causative agents. In another published study, the most common bacteria were S. pneumoniae and S. aureus (18) which was almost similar to the results of our study. Regarding the study conducted in the United States, the most common bacteria isolated from the neonates were Type B- H. Influenza (45%), S. pneumoniae (18%) and N. meningitides (14%) (18); while another similar research reported that in 70% of the 1-5 year- old children E. coli, Beta- hemolytic Streptococci, H. Influenza, N. meningitides and S. pneumoniae were screened (19). Laxer reported that, S. pneumoniae was identified as the main meningitis causative agent in children (20). In the study of Youssefi, the results of antibiogram of organisms separated from meningitis patients showed that Gram positive bacteria, the S. pneumoniae and Alfa- hemolytic Streptococci revealed good sensitivity to many of the tested antibiotics especially the antibiotics of Aminoglycosides family such as kanamycin and gentamycin, as well as the antibiotics of cephalosporin family such as cephalexin and cefotaxime; however, they showed relatively high resistance to the antibiotics of Beta Lactam family including amoxicillin and ampicillin. On the other hand, the Gram positive bacteria including S. aureus and S. epidermidis showed relatively higher resistance than most antibiotics used in this study; and their highest sensitivity was against the cminoglycosides (canamycin and centamycin) (21). In our study, coagulase –negative Staphylococci were completely sensitivity to cefixime, cefotaxime, ciprofloxacin and gentamycin; on the other hand, it was 100% resistant to methicillin. S. pneumoniae was sensitive to ceftazidime, ciprofloxacin and gentamycin, while it was completely resistant to methicillin.
In the present study; amikacin showed high sensitivity for E. coli, S. epidermidis, P. aeruginosa and S. aureus related infections. Given the findings and comparing them with published paper showed that the antibiotic resistance exists in the strains of E. coli separated from urinary tract infections; therefore, using new drugs such as Imipenem is recommended (22).
Previously published studies revealed that, the most common bacteria in the blood culture were Coagulase- negative Staphylococci, Klebsiella spp., and E. coli (23). In addition, in a conducted study in India (2015), the Gram positive bacteria were the most common agents (65.8%) and among those bacteria, Coagulase- negative Staphylococci (88.5%) were the most prevalent strains (24). In the present study, the most common bacteria in the blood specimens were Coagulase- negative Staphylococci, S. epidermidis and K. pneumoniae; respectively, which are in agreement with the previous studies. Hemmati reported that, the more prevalent Gram Positive bacteria extracted from the blood specimens of hospitalized patients in ICU, were Coagulase- negative Staphylococci (35.6%) and S. aureus (21.8%); and the most common Gram Negative bacteria were E. coli (19.8%) and K. pneumoniae (10.9%) (25).
Conclusion
Regarding the bacterial isolates from the target hospital wards and in view of the antibiotic resistance pattern as assessed in strains, our study indicated that the rates of bacterial meningitis and bacteremia in mentioned hospitals were high. Therefore, the elimin-ation and control of bacteremia and bacterial menin-gitis requires a fundamental attention. Resistance to selected antibiotics as the treatment tools could be life threatening, so it is necessary to evaluate and select effective antibiotics for etiological agents. Finally, designing an efficient method for eliminating bacteria in the hospitals and selecting a suitable medical method with a suitable antibiotic regimen should be taken into close consideration.
Restrictions:
Responsible genes to antibiotic resistance and genetic relationship between the resistant strains are not determined and these are the limitations of this study. Moreover, bacterial isolates were basically identified by biochemical features.
Acknowledgments
The authors wish to acknowledge Mazandaran University of Medical Sciences.
Conflicts of Interest
The authors declare that they have no competing interests.
Received: 2021/01/27 | Accepted: 2021/04/13 | Published: 2022/01/31
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |



